Vol. XXXI Issue 1
Article 5
ARTÍCULOS ORIGINALES
Alternatively polyadenylated calpastatin transcripts in bovine muscles
Transcriptos alternativamente poliadenilados de calpastatina en músculos de bovino
Casale M.F.1, Silvestro C.1, Corva P.M.2, Soria L.A.1
1 Universidad de Buenos Aires,
Facultad de Ciencias Veterinarias,
Cátedra de Genética,
Buenos Aires, Argentina.
2 Universidad Nacional de Mar
del Plata, Facultad de Ciencias
Agrarias, Departamento
de Producción Animal, Buenos
Aires, Argentina.
Corresponding author:
Liliana A. Soria
lsoria@fvet.uba.ar
DOI: 10.35407/bag.2020.31.01.05
Received: 09/30/2019
Revised version received: 03/05/2020
Accepted: 04/30/2020
ABSTRACT
Calpastatin activity has a key role in the tenderization process that occurs during postmortem storage of meat under refrigerated conditioning. The regulation of calpastatin (CAST) expression is highly complex, the gene has four putative promoters and at least three different polyadenylation sites, and it is also alternatively spliced. We investigated the presence of alternative polyadenylation (APA) isoforms of CAST transcripts in three muscles (infraspinatus, triceps brachii and semitendinosus) of two bovine breeds (Angus and Brahman). The 3´ RACE-PCR was used to specifically amplify the different APA sites. The amplified fragments were cloned and sequenced. Sequencing confirmed the existence of three expected polyadenylation sites corresponding to short, medium and long polyadenylated transcripts. Also, transcripts with a novel APA site were found in the three muscles of both breeds. Because the same APAs isoforms were found between muscles and breeds, we could hypothesize a possible contribution to the relative abundance of different isoforms, probably in coordination with promoter preference and alternative splicing. This knowledge would be useful in the design of future experiments to analyze differential expression of CAST isoforms and their contribution to the definition of beef tenderness.
Key words: Beef cattle; Alternative polyadenylation; 3´ RACE-PCR.
RESUMEN
La actividad de la calpastatina tiene un rol clave en el proceso de tiernización postmortem de la carne durante su almacenamiento refrigerado. La regulación de la expresión de calpastatina (CAST) es altamente compleja; el gen tiene cuatro potenciales promotores, diferentes sitios de poliadenilación de transcriptos y también splicing alternativo. En este trabajo se investiga la presencia de isoformas de transcriptos de CAST alternativamente poliadenilados (APA) en tres músculos (infraspinatus, triceps brachii y semitendinosus) de dos razas bovinas (Angus y Brahman). Se utilizó la técnica de 3´ RACE-PCR para amplificar específicamente los diferentes sitios APA. Los fragmentos amplificados fueron clonados y secuenciados. La secuenciación confirmó la existencia de tres sitios de poliadenilación conocidos. Un nuevo sitio APA fue identificado en transcriptos de los tres músculos y en ambas razas. Dado que cualitativamente no hubo variación en la presencia de isoformas definidas por APA entre músculos y razas de terneza contrastante, podría hipotetizarse una posible contribución a la abundancia relativa de distintas isoformas, probablemente en forma coordinada con la elección de promotores y el splicing alternativo. Este nuevo conocimiento podría ser de utilidad para el diseño de experimentos de análisis de expresión diferencial de isoformas de calpastatina, para ponderar la contribución de las mismas a las variaciones en terneza de la carne.
Palabras clave: Bovinos para carne; Poliadenilación alternativa; 3´ RACE-PCR.
INTRODUCTION
The calpain/calpastatin system is an endogenous,
calcium-dependent proteinase system. Calpain is
involved in the breakdown of protein myofibrils;
calpastatin inhibits calpain activity and, therefore,
partially regulates postmortem proteolysis in muscle
(Koohmaraie et al., 1996). This enzyme complex
affects some meat quality traits; particularly it plays an
important role in meat tenderization (Shackelford et al.,
1995). Bos indicus breeds (e.g. Brahman) are well known
for their higher calpastatin activity in muscle, which
inhibits protein degradation and results in tougher beef
(Whipple et al., 1990; Pringle et al., 1997).
The bovine calpastatin (CAST) gene consists of 35
exons spanning at least 130 kb on chromosome 7 (Bishop
et al., 1993; Raynaud et al., 2005a). Four alternative
promoters direct the expression of four different
transcripts isolated from different tissues, named Type
I, II, III, and IV, which differ in their 5´ ends (Raynaud
et al., 2005a). Moreover, differences in transcript length
can also be originated by alternative polyadenylation
sites and alternative exon splicing (Cong et al., 1998;
Raynaud et al., 2005b; Nattrass et al., 2014).
The polyadenylation (poly A) reaction of mammalian
pre-mRNAs proceeds in two stages: first the cleavage of
pre-mRNA and then the addition of poly(A) tail to the
newly formed 3´ end. Polyadenylation is important for
translation efficiency, stability, and cellular localization
of mature mRNA (Elkon et al., 2013). Many eukaryotic
genes contain more than one polyA (pA) site, leading
to the generation of distinct mRNA isoforms from the
same gene through alternative polyadenylation (APA)
(Tian et al., 2017). The APA sites of CAST are located in
the 3′ untranslated region (3′UTR), leading to alternative
transcripts, all with the same coding frame but with
variable 3′UTRs (called UTR-APA). It should be noted
that although the UTR-APA isoforms do not affect the
coding frame, they might lead to changes in mRNA halflife
or translation efficiency, since longer 3′UTRs can
have more microRNA binding sites, more RNA-binding
protein recognition sites, or altered RNA secondary
structure (Millevoi and Vagner, 2010; Mayr, 2016).
Three polyadenylated variants in the 3´ UTR have
been described for bovine CAST transcripts, named:
short, medium and long (Cong et al., 1998; Raynaud et
al., 2005b).
The medium form is 789 bp longer than the short one,
whereas medium and long forms differ in 1089 bp. Until
now, there is no conclusive information about potential
associations between promoter use and alternative
polyadenylation sites. However, the type III isoform
seems to be expressed in combination with all the three
reported 3´ UTRs (Raynaud et al., 2005b).
A potential association between the relative
abundance of CAST isoforms and beef tenderness
has been reported. Not only breed differences in beef
tenderness but also among muscles of the same breed
have been extensively documented (Rhee et al., 2004;
Calkins and Sullivan, 2007). Therefore, we took muscle
samples of three muscles: infraspinatus (more tender),
triceps brachii and semitendinosus (tougher), from a Bos
taurus breed (Angus) and a Bos indicus breed (Brahman)
that is known to produce consistently tougher beef
compared to European breeds.
The objective of this study was to analyze the presence
of APA variants of CAST in samples of the muscles and
breeds mentioned above.
For this purpose we used the method known as “Rapid
Amplification of 3´-cDNA End” (3´ RACE) (Frohman
et al., 1988) and sequencing to detect and characterize
transcripts that differ in their 3´ UTR length.
MATERIALS AND METHODS
Samples
Within 1 h after slaughter, muscle samples (2 g)
were taken from infraspinatus, triceps brachii and
semitendinosus of 2 Angus steers (364±17 kg final body
live weight and 19 months of age on average) and 2
Brahman steers (408±12 kg final body live weight and 42
months of age on average) and stored in liquid nitrogen
or at 4 ºC as needed. The steers were slaughtered at two
local private abattoirs: Carnes del Salado SA (Castelli,
Buenos Aires, Argentina) and Don Rafael SRL (Santo
Tomé, Corrientes, Argentina) for Angus and Brahman
respectively. The animals were slaughtered after a
24 h rest in paddocks without feed but with access to
water, according to the Handbook of Procedures for
Animal Welfare of the National Service for Animal
Health (Servicio Nacional de Sanidad Animal, SENASA) of
Argentina.
RNA isolation
Approximately 100 mg of muscle tissue pulverized
in a small amount of liquid nitrogen with a cooled
pestle were mixed in 1 mL of TRIzol reagent (Life
Technologies Corporation, CA, USA) and homogenized
with the help of a mixer (Velp Scientifica®, Usmate,
MB, Italy). The supernatant (aqueous phase containing
the RNA) of each homogenate was obtained according
to the manufacturer’s protocol. The aqueous phase
was mixed with ethanol (Sigma-Aldrich, St Louis, MO,
USA) and total RNA was purified with RNA Clean &
Concentrator®-5 kit (Zymo Research, Irvine, CA, USA)
as per the manufacturer’s instructions. RNA quality
and concentration were determined by the OD260/280
value (> 1.7) with a NanoDrop Lite Spectrophotometer
(Thermo Fisher Scientific, Waltham, MA, USA) and
confirmed by electrophoresis in agarose gels (0.8%)
stained with GelRed® Nucleid Acid Gel Stain (Biotium,
Fremont, CA, USA).
Reverse transcription -3´ rapid amplification of cDNA
ends-PCR
The 3´ rapid amplification of cDNA ends or 3´ RACE
(Frohman et al., 1988) is widely used to isolate the
cDNA of unknown 3´ flanking sequences. The 3´ RACE
technique was used to specifically amplify the different
polyadenylation sites of CAST transcripts (Figure 1).
The oligodT used to generate the cDNA included an
adapter sequence that was complementary to the CAST antisense primer (Table 1 and Figure 1). Two μg of RNA
were used to produce first strand cDNA using 200 U
of M.MLV Reverse transcriptase enzyme (Promega,
Madison, WT, USA), 40 U of Recombinant Rnasin
(Promega, Madison, WT, USA), 5 mM of DTT (Promega,
Madison, WT, USA), 2.5 uM of oligodT-adapter (Table 1)
and 10 pmoles of dNTPs (Promega, Madison, WT, USA).
Two specific sense oligonucleotides (Cast-e28
and Cast-e30) (Table 1) were designed to ensure the
amplification of 3´UTRs of different length, as described
by Raynaud et al. (2005b) and also predicted with Poly
(A) Signal Miner software (Liu et al., 2003).
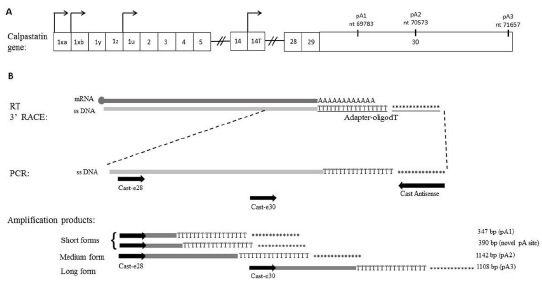
Figure 1. A. Schematic representation of calpastatin gene showing the four transcription start sites, exons and
previously described polyadenylation sites (indicated as pA1, pA2 and pA3). B. Scheme of reverse transcription
3´RACE-PCR technique with the four amplification products obtained. “********” corresponds to sequence
GATACGCCGCGATTCGAACCTGACCATGTACAGCTGCCC. Cast antisense primer is complementary to the adapter-oligodT.
The PCR products are aligned with their corresponding forward primer (Cast-e28 or Cast-e30).
Table 1. Oligonucleotide sequences used for cDNA synthesis (3´ RACEPCR).

The different end-point PCR reactions were performed with 3μL of cDNA, 10 pmoles of antisense oligonucleotide (Cast-antisense) which hybridizes to the adapter region of the adapter-oligodT (Table 1), 10 pmoles of the corresponding sense primer (Cast-e28 or Cast-e30), 2 U of Platinum Taq Polymerase (Invitrogen, São Paulo, Brazil), 2mM of Cl2Mg and 10 pmoles of dNTPs. Cycling conditions were 94 °C 5 min, 35 cycles of 94 °C for 1 min, 60 °C for 30 sec and 72 °C for 1 min; followed by a final 2 min extension at 72 °C. Since there was no certainty about any preferences in polyA sites among muscles or breeds, all the obtained amplicons were considered. However, special attention received the amplicons of approximately 300 bp and 1100 bp (primer Cast-e28) and 1055 bp (primer Cast-e30) which corresponds to previously described APA variants (Raynaud et al., 2005b). Amplified PCR products were observed by 1.5 % agarose gel electrophoresis with GelRed® Nucleid Acid Gel Stain (Biotium, Fremont, CA, USA). Selected bands were eluted with the PureLink Quick Gel Extraction and PCR Purification Combo Kit (Invitrogen, Carlsbad, CA, USA) and cloned in the pGEM-T-easy system (Promega, Madison, WT, USA) and Escherichia coli DH5alfa competent cells according to the manufacturer´s protocol. Forward and reverse sequences (using Sp6 and T7 primers) were generated from each cloned amplicon in an Applied Biosystems 3100 DNA Sequencer. These sequences were then comparatively analyzed by BLASTn and aligned to the reference genomic sequence of calpastatin (71,657 bp, Genebank accession AH014526.2) for the identification of the different polyadenylation sites.
RESULTS
The presence of alternatively polyadenylated calpastatin
transcripts in three bovine muscles (infraspinatus, triceps
brachii and semitendinosus) from two cattle breeds
(Angus and Brahman) was analyzed.
The 3´RACE-PCR system was designed in order to
obtain amplicons of approximately 350 bp and 1200 bp
for short and medium transcripts with the Cast-e28
oligonucleotide; and 1100 bp for the long polyadenylated
transcript using with the Cast-e30 oligonucleotide. In
the PCR that included the forward primer Cast-e28,
four intense bands (approximately 350, 400, 800 and
1200 bp) were observed on the agarose gel, whereas in
sample no. 6 a product of approximately 1300 bp was
also amplified (Figure 2A). Figure 2B shows two intense
bands (800 and 1100 bp) obtained by PCR amplification
with forward primer Cast-e30. All these bands were
eluted, cloned and sequenced to confirm their identity.
Positive clones of 347, 390, 1108 and 1142 bp were
obtained (Figure 1). Sequencing confirmed the existence
of three expected polyadenylation sites at positions
69783, 70573 and 71657 of the CAST reference sequence
(GenBank accesion AH014526.2) corresponding to
short (347 bp) and medium (1142 bp) polyadenylated
transcripts amplified with Cast-e28; and long (1108 bp)
polyadenylated transcripts amplified with Cast-e30
(Figure 3). These results are in agreement with the
APA sites reported by Cong et al. (1998), Raynaud et al.
(2005b) and Natrass et al. (2014). Interestingly, PCR
performed with Cast-e28 produced a novel APA site
that was present in the three muscles of both breeds.
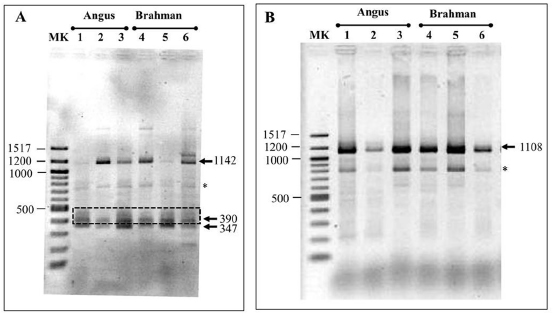
Figure 2. 1% agarose gel electrophoresis showing alternatively polyadenylated CAST transcripts amplified by 3´ RACE-PCR,
using forward primers Cast-e28 (A) or Cast-e30 (B). Staining was performed with GelRed® Nucleid Acid Gel Stain (Biotium,
Fremont, CA, USA). Each line was loaded with 10 μl of the PCR.
Lanes 1 and 4: Infraspinatus, 2 and 5: Triceps brachii, 3 and 6: Semitendinosus. MK: Quick-Load 100 bp DNA ladder (New
England BioLabs, Hitchin, UK). Spurious fragments are indicated with a star (*). The amplicon corresponding to a novel pA site
is indicated with a box.
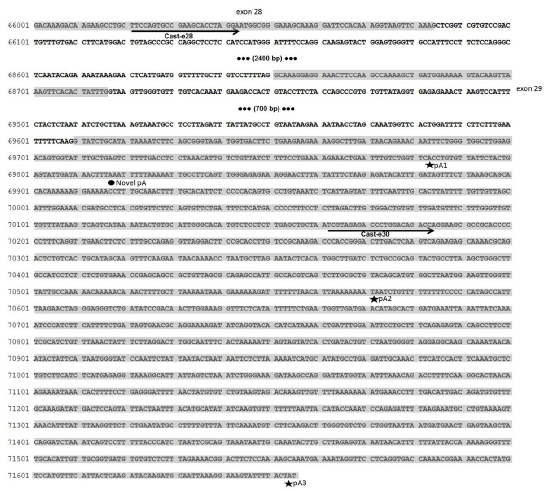
Figure 3. Sequence of the distal region of the calpastatin gene (Exons 28 to 30). Numbers on the left correspond to
coordinates of reference sequence AH014526.2. Exons 28, 29 and 30 are shadowed. Non relevant parts of intron were
removed and its base pairs annotated between brackets. Primers Cast-e28 and Cast-e30 are indicated with arrows. Known
polyadenylation signals (pA1, pA2 and pA3) are indicated with a star (); a circle () indicates a novel polyadenylation site.
This APA site is located in position 69817 of the reference sequence AH014526.2 (Figure 3) and corresponds to a 390 bp amplicon (Figure 2). Sequencing determined that the 800 bp (Figure 2A and B) and 1300 bp (Figure 2A, lane 6) bands corresponded to a spurious amplicon. The expression of the four different polyadenylated mRNA isoforms was confirmed in the 12 samples included in this study (Figure 2) and in all the samples that were analyzed for the implementation of the methodology (data not shown).
DISCUSSION
There is still little information about the expression of
CAST isoforms with different polyadenylation sites in
different muscles or breeds, and its potential effects
on beef quality traits. Nattrass et al. (2014) quantified
two polyadenylation variants of CAST (those designated
here as short and long, respectively) in the longissimus
lumborum muscle of Angus and Brahman steers. The
steers had been genotyped for the CAST:c.2832 A>G SNP,
one of the first genetic markers for beef tenderness to be
commercially available (Barendse, 2002). The findings
of that study showed that a lower concentration of
mRNA terminating at the proximal site (short) was
significantly associated with the favorable allele for
beef tenderness (A). These results supported the
conclusion that CAST:c.2832 A>G SNP may be in linkage
disequilibrium with regulatory sequences which have a role in the post-transcriptional processing of CAST
transcripts, leading to reduced levels of calpastatin
protein in muscles of individuals carrying the favorable
allele. No association between CAST:c.2832 A>G SNP and
polyadenylated forms were found in our samples since
all the samples tested were homozygous for the A allele
(data not shown).
A general correlation between the level of gene
expression and the relative abundance of 3´ UTR
isoforms have been reported (Ji et al., 2011). The
correspondence between APA and gene expression may
be the consequence of the coupled usage of alternative
promoters and polyA sites, previously reported for some
genes (Costessi et al., 2006; Winter et al., 2007). Since
mRNAs with short 3´UTRs are generally more stable
due to avoidance of destabilizing elements binding
to that region (Mayr and Bartel, 2009) and the escape
from cellular mechanisms degrading long 3´UTRs (Hogg
and Goff, 2010), a comparatively higher expression
of short 3´UTR isoforms would lead to a higher steady
concentration of mRNA, and the opposite would also
hold true.
The results reported here did not allow us to establish
a connection between alternative polyadenylation of CAST and calpastatin activity. However, they show that
alternative polyadenylation, probably coupled with the
usage of alternative promoters, adds more complexity
to the analysis of beef tenderization both between
breeds and between muscles within a breed. According
to Raynaud et al. (2005b), the type III isoform is the most
abundantly expressed transcript in muscle, and it would
also present polyadenylation variants.
Three known APA transcripts and a novel isoform of
the bovine CAST gene were identified in three muscles
that differ in tenderness, of two cattle breeds with
known variation in calpastatin activity. Thus, variability
in beef tenderness does not seem to be simply due to
the presence or absence of a given APA form. Moreover,
the biological significance and implications for muscle
physiology of at least four alternative polyA sites are not
well understood.
New research would be needed for the relative
quantification of each isoform and the evaluation of
their effects on calpastatin activity and ultimately,
on beef tenderness. All known APA isoforms should
be comparatively quantified in order to get a better
understanding of the contribution of CAST expression
to the variability in beef tenderness, both among breeds
and muscles within a breed. The knowledge of the new
isoform would help in the design of qPCR experiments
and reinforces the concept that a complex gene such as
CAST should be also evaluated through full length RNA
sequencing to detect the potential associations between
APA, alternative splicing and alternative promoter
selection.
ACKNOWLEDGEMENTS
Research supported by the National Agency of Science and Technology (ANPCyT) of Argentina, Grant PICT 2014-1298. Angus and Brahman steers were provided by the Integrated Experimental Farm (Chacra Experimental Chascomús, Argentina) and a private beef cattle ranch (Don José, Virasoro, Corrientes, Argentina) respectively.
BIBLIOGRAPHY
1. Barendse W.J. (2002) DNA markers for meat tenderness. International Patent Publication W0 02/064820.
2. Bishop M.D., Koohmaraie M., Killefer J., Kappes S. (1993) Rapid communication: restriction fragment length polymorphisms in the bovine calpastatin gene. J. Anim. Sci. 71 (8): 2277.
3. Calkins C.R., Sullivan G. (2007) Ranking of beef muscles for tenderness. Beef Research in www.beefresearch.org›CMDocs›PE_Fact_Sheets (accessed March 2020).
4. Cong M., Thompson V.F., Goll D.E., Antin P.B. (1998) The bovine calpastatin gene promoter and a new N-terminal region of the protein are targets for cAMP-dependent protein kinase activity. J. Biol. Chem. 273 (1): 660-666.
5. Costessi L., Devescovi G., Baralle F.E., Muro A.F. (2006) Brain-specific promoter and polyadenylation sites of the beta-adducin pre-mRNA generate an unusually long 3′-UTR. Nucleic Acids Res. 34: 243-253.
6. Elkon R., Ugalde A.P., Agami R. (2013) Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14 (7): 496-506.
7. Frohman M.A., Dush M.K., Martin G.R. (1988) Rapid production of fulllength cDNAs from rare transcripts: Amplification using a single genespecific oligonucleotide primer. Proc. Natl. Acad. Sci. 85: 8998-9002.
8. Hogg J.R., Goff S.P. (2010) Upf1 senses 3´UTR length to potentiate mRNA decay. Cell 143 (3): 379-389.
9. Ji Z., Luo W., Li W., Hoque M., Pan Z., Zhao Y., Tian B. (2011) Transcriptional activity regulates alternative cleavage and polyadenylation. Mol. Syst. Biol. 7: 534.
10. Koohmaraie M. (1996) Biochemical factors regulating the toughening and tenderization process of meat. Meat Sci. 43: 193-201.
11. Liu H., Han H., Li J., Wong L. (2003) An Insilico method for prediction of polyadenylation signals in human sequences. Proceedings of 14th International Conference on Genome Informatics (GIW 2003), pp. 84-93.
12. Mayr C., Bartel D.P. (2009) Widespread shortening of 3´UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138 (4): 673-684.
13. Mayr C. (2016) Evolution and Biological Roles of Alternative 3´UTRs. Trends Cell Biol. 26 (3): 227-237.
14. Millevoi S., Vagner S. (2010) Molecular mechanisms of eukaryotic premRNA 3´ end processing regulation. Nucleic Acids Res. 38 (9): 2757- 2774.
15. Nattrass G.S., Café L.M., McIntyre B.L., Gardner G.E., McGilchrist P., Robinson D.L., Wang Y.H., Pethick D.W., Greenwood P.L. (2014) A post-transcriptional mechanism regulates calpastatin expression in bovine skeletal muscle. J. Anim. Sci. 92 (2): 443-455.
16. Pringle T.D., Williams S.E., Lamb B.S., Johnson D.D., West R.L. (1997) Carcass characteristics, the calpain proteinase system, and aged tenderness of Angus and Brahman crossbred steers. J. Anim. Sci. 75 (11): 2955-2961.
17. Raynaud P., Jayat Vignoles C., Laforêt M.P., Levéziel H., Amarger V. (2005a) Four promoters direct expression of the calpastatin gene. Arch. Biochem. Biophys. 437 (1): 69-77.
18. Raynaud P., Gillard M., Parr T., Bardsley R., Amarger V., Levéziel H. (2005b) Correlation between bovine calpastatin mRNA transcripts and protein isoforms. Arch. Biochem. Biophys. 440 (1): 46-53.
19. Rhee M.S., Wheeler T.L., Shackelford S.D., Koohmaraie M. (2004) Variation in palatability and biochemical traits within and among eleven beef muscles. J. Anim. Sci. 82 (2): 534-550. doi:10.2527/2004.822534x.
20. Shackelford S.D., Wheeler T.L., Koohmaraie M. (1995) Relationship between shear force and trained sensory panel tenderness ratings of 10 major muscles from Bos indicus and Bos taurus cattle. J. Anim. Sci. 73: 3333-3340.
21. Tian B., Manley J.L. (2017) Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell. Biol. 18 (1): 18-30.
22. Winter J., Kunath M., Roepcke S., Krause S., Schneider R., Schweiger S. (2007) Alternative polyadenylation signals and promoters act in concert to control tissue-specific expression of the Opitz Syndrome gene MID1. BMC Mol. Biol. 8: 105.
23. Whipple G., Koohmahraie M., Dikeman M.E., Crouse J.D., Hunt M.C., Klemm R.D. (1990) Evaluation of attributes that affect longissimus muscle tenderness in Bos taurus and Bos indicus cattle. J. Anim. Sci. 68 (9): 2716-2728.



