Vol. XXXIII Issue 2
Article 2
DOI: 10.35407/bag.2022.33.02.02
ARTÍCULOS ORIGINALES
Impact of genetic ancestry on the distribution of interferon-λ4 rs12979860 polymorphism in a global population of Buenos Aires,
Argentina
Impacto de la
ancestría genética en la distribución del polimorfismo de interferón-λ4
rs12979860 en una población global de Buenos Aires, Argentina
Mansilla F.C.1 *
Avena S.A.2,3,4
Dejean C.B.2,3
Turco C.S.1
Capozzo A.V.1,4
1 Instituto de Virología e Innovaciones Tecnológicas
(IVIT), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) -
Instituto Nacional de Tecnología Agropecuaria (INTA), Hurlingham, Buenos Aires,
Argentina.
2 Centro de Ciencias Naturales, Ambientales
y Antropológicas (CCNAA), Universidad Maimónides, Buenos Aires, Argentina.
3 Sección Antropología Biológica,
Instituto de Ciencias Antropológicas (ICA), Facultad de Filosofía y Letras, Universidad
de Buenos Aires, Buenos Aires, Argentina
4 Consejo Nacional de Investigaciones Científicas
y Técnicas (CONICET), Argentina.
* Corresponding author: Mansilla F.C. mansilla.florencia@inta.gob.ar ORCID 0000-0002-8325-0579
ABSTRACT
Human interferon-λ4 is a cytokine involved in early stages of antiviral responses.
Strikingly, some allelic variants with
diminished antiviral activity reduce the susceptibility to viral infections,
thus they would have suffered a
positive selection pressure throughout the evolutionary history of the genus Homo. An intronic variant
within the IFNλ4 locus (rs12979860, T˃C) emerged as one of the main gene determinants of the response to
HCV and other viruses. The rs12979860-C allele has a differential frequency in African,
European and Native American populations, though South American data are scarce. Here we
characterize for the first time the distribution of rs12979860 genotypes in a sample of the global
population of Buenos Aires, Argentina, assessing its association with European, Native American
and African parental components. The rs12979860 genotypes were determined by PCR-RFLP in DNA
samples from donors of a blood banks of Buenos
Aires (n=96), whose genetic individual ancestry (European, African or Native
American) had been previously
determined using molecular markers. The distribution of rs12979860-CC, CT
and TT was 29.17%, 50.0% and 20.83%,
respectively. A significant increase in the frequency of CC among donors with a strong European contribution
and a greater impact of the Native American
component among donors carrying the T allele were observed. Native
American and European components were
associated to the rs12979860 distribution in a sample of the global population
of Buenos Aires, while no differences
were directly attributable to the African ancestry. Considering interferon´s key role in
antiviral responses, our results may contribute to both bioanthropological and immunogenetic studies associated with
infectious diseases.
Key words: Ancestry, Buenos Aires, IFNλ4 polymorphism, rs12979860
distribution.
RESUMEN
El
interferón-λ4 humano es una citoquina involucrada en la respuesta
antiviral. Algunas variantes alélicas
con menor actividad antiviral, paradójicamente, reducen la susceptibilidad a
infecciones virales, por lo que habrían
sufrido una presión de selección positiva en la historia evolutiva del gיnero Homo.
Una variante dentro del locus de IFNλ4 (rs12979860, T˃C), con
distribución diferencial en poblaciones
africanas, europeas y nativas americanas, surgió como uno de los principales determinantes genéticos de la
respuesta al HCV y otros virus. Aquí caracterizamos por primera vez la distribución de los
genotipos de rs12979860 en una muestra de la población cosmopolita de Buenos Aires, Argentina,
evaluando el impacto de su ancesrtría. Se determinaron diferentes genotipos de rs12979860 por
PCR-RFLP en muestras de ADN de donantes de bancos de sangre de Buenos Aires (n=96), cuya
ancestría individual había sido previamente determinada mediante diferentes marcadores moleculares.
La distribución global de rs12979860-CC, CT
y TT fue 29,17%; 50,0% y 20,83%, respectivamente. Se observó un aumento
significativo de la frecuencia del
genotipo CC entre individuos con fuerte aporte europeo y un mayor impacto
del componente nativo-americano entre
portadores del alelo T. Los componentes nativo-americano y europeo se asociaron a la distribución
rs12979860 en una muestra poblacional global de Buenos Aires, mientras que no se vieron diferencias
directamente asociadas a la ancestría africana. Considerando el papel clave del interferón
en la respuesta antiviral, nuestros resultados pueden contribuir a estudios con un enfoque
bioantropológico así como a estudios inmunogenéticos asociados a enfermedades infecciosas.
Palabras clave: Ancestría,
Buenos Aires, Polimorfismo en
IFNλ4, Distribución de rs12979860.
Received: 03/04/2022
Revised version received: 06/30/2022
Accepted: 08/08/2022
General Editor: Elsa Camadro
INTRODUCTION
Lambda interferons (IFNλ) are cytokines rapidly produced
by most vertebrates during the innate immune
response, constituting the first line of defense against viral infections (Lazear et al., 2015). IFNλ1, 2 and 3 were identified in
2003 (Kotenko et al., 2003; Sheppard et al., 2003) and in 2013 a functional form of IFNλ4 was firstly characterized (Prokunina-Olsson et
al., 2013). The IFNλ4 locus (19q13.2) is highly polymorphic (Fang et al.,
2020) and it was reported
that some allelic variants can modulate
the susceptibility, progression and response to treatments against different viral
infections (Chatterjee, 2010; Bravo et al., 2014; Angulo et al., 2015; Ispirologlu et al., 2017; da Silva Cezar et al., 2020). Interestingly, the most
favorable alleles in this regard correspond to
mutations that are in strong linkage disequilibrium and restrict the expression, stability or
antiviral activity of IFNλ4 (Booth and George, 2013; O’Brien et al., 2014; Prokunina-Olsson, 2019).
Throughout the evolutionary
history of the genus Homo these
mutations have suffered a positive selection
pressure resulting in a differential global distribution which is correlated to the ancestry of
different human populations and may
affect the immune response to different
pathogens (Key et al., 2014; Bamford et al., 2018). An intronic variant that reduces the antiviral activity of IFNλ4 (rs12979860, T˃C) was characterized as the main gene determinant of the response
against Hepatitis C Virus (HCV). The
rs12979860-T allele is associated with
lower sustained virologic response
(SVR) rates and a lower percentage of treatment success (Ge et al., 2009). On the
other hand, the CC genotype was
strongly associated with spontaneous
resolution and lower susceptibility to HCV infection (Thomas et al., 2009; Pedergnana et al., 2012; Indolfi et al., 2014a; Fan et al., 2016). Moreover, genotyping of
rs12979860 is recommended to predict the patient´s response to different antiviral treatments (Sharafi et al., 2012; Ramamurthy et al., 2018). Different correlations
between rs12979860 and clinical phenotypes associated with other viral infections have also been
reported, conditioning the
susceptibility, evolution and/or
response to treatment against Hepatitis B and D (Ispirologlu et al., 2017), Dengue (da Silva Cezar et al.,
2020), HIV (Chatterjee, 2010; Zaidane et al., 2018), CMV (Bravo et al., 2014; Chmelova et al., 2019) and coronaviruses (Hamming et al.,
2013).
The rs12979860-C allele has a
global frequency of 0.23-0.55 in
African populations; 0.53-0.80 for
Europeans and 0.72-1.00 for Asians, with higher frequencies in eastern Asia. Data about the
distribution of these variants in South
American populations are scarce and tend
to be biased due to the small sample size
and the genetic admixture of the populations assessed. The Argentinean population´s ancestry is the
result of a deep miscegenation, product
of different migratory waves during the
last centuries, which means that the
European, Native American and African components (frequently underestimated) are present at
different degrees in the gene pool of
different cosmopolitan populations of
the country (Avena et al., 2012). In this regard, the
immunogenetic profiling of IFNλ4- rs12979860, and the association
with its ancestry, may be a potential
tool in both anthropological and biomedical
studies associated with infectious diseases. The objective of this study was to determine the
distribution of the allelic variants of
rs12979860 in a cosmopolitan population
of Buenos Aires, Argentina, whose ancestry
had been previously determined by assessing a set of 106 biallelic SNPs (Ancestry Informative
Markers) widely spaced and balanced
throughout the genome, that can
discriminate Native American, African and European ancestry (Avena et al., 2012).
MATERIALS AND METHODS
Study Design
This study comprised DNA samples
from unrelated donors from both public
and private hospitals blood banks in
Buenos Aires, Argentina (n=96). Informed
consent was obtained from all individual participants included in the study. Most of them (89/96) also
agreed to provide information about the
region/country of birth of all their
grandparents, which was included in the
data analysis. The study was approved by the Ethics Committee of the Hospital Italiano of Buenos
Aires and was performed in accordance
with the ethical standards adopted in
the Declaration of Helsinki.
rs12979860 genotyping
Different genotypes of rs12979860
were determined by PCR-RFLP, as it was
previously described (Sharafi et al., 2012). A 241 bp fragment was amplified by
endpoint PCR (Taq Pegasus®, Productos Bio-Lógicos, Bs. As., Argentina) following a standard
cycle (5 min at 94° C; 35 cycles of 20
s at 94° C, 20 s at 59° C and 20 s at
72° C; and 5 min at 72° C) and then digested
with Bsh12361 restriction enzyme (Thermo Fisher, DE, USA; 1U/reaction) for 1 h at 37° C. The
primers used were
5´GCGGAAGGAGCAGTTGCGCT3´ (Fw) and
5´TCTCCTCCCCAAGTCAGGCAACC3´ (Rv) and the resulting fragments (rs12979860-CC = 196 +
45 bp; rs12979860-CT = 241 + 196 + 45
bp; rs12979860-TT = 241 bp) were
revealed by agarose gel electrophoresis
(3%) stained with GelRed (Biotium, CA, USA).
Statistical analysis
The allelic frequencies were
determined, and Hardy- Weinberg
equilibrium was assessed using the chisquare
test (Microsoft Excel GenAIEx 6.5, Peakall and
Smouse, 2012) to compare the
genotype distribution. Differences
associated to European, Native American
or African component were determined using T test (GraphPad Prism 9). In all statistical
analysis a p<0.05 was
considered as statistically significant and α=0.05 was set as the risk
level.
RESULTS AND DISCUSSION
The average individual ancestry
was estimated as 69.4% European, 26.3%
Native American and 4.3% African.
Frequencies lower than 0.02 were not included in the data analysis since they may be associated
to technical artifacts. The European
component was present in every tested
sample, with individual frequencies ranging from 0.02 to 1. The Native American component was
also detected but to a lesser extent,
in 79% of the samples (frequencies
0.02-0.8). Finally, the African ancestry
was detected in 41% of the samples, with a frequency range from 0.02 to 0.23 (Figure 1, modified from Avena et al., 2012). This evidences the multiplicity of origins of Buenos Aires´ population, resulting of the
miscegenation between Native Americans,
enslaved Africans who came mainly from
West Africa and Mozambique until the
first half of the 19th century (Fejerman et al., 2005) and European immigrants,
mainly from Italy and Spain, who
arrived in the country between 1870 and 1960 (Avena et al., 2006; Muzzio et al., 2018). These results are in line
with previously published data (Avena et al., 2006), further challenging the European
self-perception as Argentina’s
identity.

Figure 1. Frequency
distribution of the individual European (a), Native American (b) and African
ancestry (c) among healthy donors from Buenos
Aires, Argentina, enrolled in this study (n=96). Modified from Avena et al. (2012).
Several studies have reported the
distribution of the rs12979860
genotypes in different populations,
mainly assessing its correlation with the susceptibility to different viral infections and response
to antiviral treatments (Wu et al.,
2012; Porto et al., 2015; Taheri et al., 2015; Echeverría et al., 2018). The correlation of this
distribution and the local ancestry of these populations as well as its implications have also been
assessed (Indolfi et al., 2014b; Rizzo et al., 2016), though this is the first
report in an Argentinean global population.
The overall distribution of rs12979860-CC, CT and TT was 29.17%, 50.0% and 20.83%, respectively.
Hardy- Weinberg equation was used to
calculate the genetic variation of this
population at equilibrium. Significant
differences were not detected (chi-square test: 0.00469; p=0.99766), thus suggesting that the
impact of posible microevolutionary
mechanisms and population structure is
not significant. The allelic frequencies for C and T were 54.17% and 45.83%, respectively.
These results differ from data reported
in HCV chronically infected patients of
a public center in Buenos Aires, with
an allelic frequency of C=0.6 and 45.0% of
heterozygosity (Machicote et al., 2018). This higher frequency of
rs12979860-C is expected as it is known
that this allele is favorable in both acute and chronic HCV infection. In this regard, the
differences observed between healthy
and infected individuals highlight the
impact of assessing global populations when studying the distribution of this kind of
markers.
A significant increase in the
frequency of CC genotype was observed
among donors with a strong European
contribution (Figure 2a, p<0.05).
Our results also suggest a greater
impact of the Native American component
among donors carrying the T allele (both
CT and TT genotypes), although differences were marginally significant (Figure 2b). No differences in the
rs12979860 distribution were directly attributable to the African component (Figure 2c, p>0.05), represented at
low levels in our sample. Based
on previously reported data on the composition
and immigration patterns of the admixed population of Buenos Aires (Avena et al., 2012), we defined our parental
population including sub-Saharan Africans (involved in slavery trafficking) and Europeans from
Italy and Spain (Avena et al.,
2006). To minimize bias, we only considered reported data on the rs12979860
distribution (Table S1) from
non-cosmopolitan populations with a sample size greater than 50. Ethiopian Jews and
Sephardic Jews from Rome, Italy, were
also excluded, as these groups tend to
be endogamous and have a different origin, which may introduce certain bias to our analysis.
The mean frequency of the rs12979860-C
allele for this parental population is
0.654 for Europeans, 0.298 for Africans
and 0.518 for Native Americans (table S1). However, data available regarding the Native American
component are scarce and are often
based either on cosmopolitan admixed
populations or studies with very small sample
sizes and variable results. Despite the lack of a robust sample to perform comparisons, our results
suggest that populations with greater
autochthonous ancestry tend to exhibit
higher frequencies of the rs12979860-T
allele. Further studies are
needed to fully characterize the
distribution of this polymorphism in Latin
America, as available data seem to be contradictory. To explain this it is important, regarding
cosmopolitan populations, to disclose
their composition and their genetic
ancestry in order to determine their parental
populations´contribution. Latin American cosmopolitan populations are known to be admixed, but the
European, Native American and
sub-Saharan contributions have marked
regional differences. Hence the relevance of
studying cases such as the one here described considering the genetic ancestry of the population under
study.
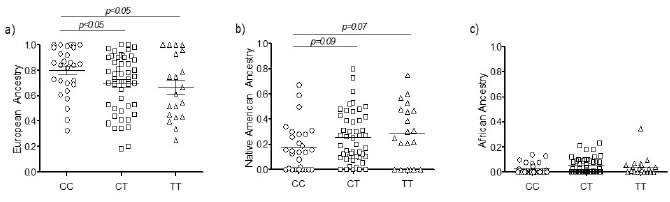
Figure 2. Distribution of
European (a), Native American (b) and African ancestry (c) among individuals
carrying rs12979860-CC, CT and TT genotypes.
p<0.05 were considered as statistically significant.
The frequency of rs12979860-C in
Buenos Aires´ individuals was similar
to previously reported data for
populations from Tuscany (C=0.603) in Italy, which are among the lowest compared to other West
European populations (Table S1). The
reported frequency of this allele in an
Iberian population, however, was higher
than the one described in our study (C=0.705, Table S1). Although immigrants from both Italy and
Spain are the main determinants of the
European ancestry of Buenos Aires´
population (Avena et al., 2006), it is to note that most of the immigrants in Buenos Aires (and
Argentina) were of Italian origin
(Avena et al. 2006). This may
explain, at least partially, the frequencies here described. In order to further characterize the
European contribution to the rs12979860
distribution we considered, when
available, the self-reported data about
grandparents’ origins. Interestingly, a total
of 53 individuals declared the nonexistence of grandparents of European origin (8/53:CC,
33/53:CT and 12/53:TT), while only 36
individuals reported at least one
grandparent from Italy, Spain/Portugal or other European countries (14/36:CC, 16/36:CT and
6/36:TT). This may be attributed to the
fact that the vast majority of immigrants
arrived in Buenos Aires before 1950. In
our sample, the presence of Iberian ancestry seems to be underrepresented, as genealogical data
suggest that the self-reported Italian
ancestry was 33.0% higher than Iberian
ancestry. Altogether, our results may be
explained by the higher presence of Italian ancestry among European descendants in our sample, as
well as by the admixture of these
individuals with Native Americans and
Africans or afro-descendants with a
higher rs12979860-T frequency, thus increasing
the heterozygosity and the rs12979860-T frequency. However, it is important to consider that,
despite being very useful especially in
regions with recent immigration
patterns (Avena et al. 2012), this kind of surveys must be carefully analyzed, since different social
and economic aspects may influence the
individual self-perceived ancestry, as
it was recently reported (Paschetta et al. 2021).
Notably, most of the populations
that have been included in large-scale
immunogenomic studies were of European origin, and might include certain bias
by demographic, social and economic
conditions of nonrandomly selected
individuals (Peng et al., 2021). This may have affected the
representativeness of the sample, thus
compromising the conclusions of those studies.
Therefore, increasing the genetic diversity while considering these structural inequalities is
mandatory in order to obtain more
reliable results. The PCR-RFLP protocol
here applied was previously described and fully validated against PCR-sequencing, with a
concordance of 100% in the results
obtained for C/T alleles (Sharafi et al., 2012). In this regard, the use of a simple low-cost and high-yielding technique is paramount, since
it allows small regional laboratories
with limited resources to conduct
population genetic studies, thus reducing the
sampling bias that may occur in large cosmopolitan cities. This is particularly relevant in
regions such as South America, in which
the availability of qPCR or sequencing
platforms is still limited. During the
last years, there has been a growing
interest on the impact of genetic ancestry on the immune response against viral infections (Mersha and
Abebe, 2015). The molecular determinants responsible for those associations are being increasingly
understood, and interferon pathways and
their expression patterns seem to be
influenced by genetic ancestry (Miretti and Beck, 2006; Randolph et al., 2021), as suggested by our
results.
In the context of the COVID-19
pandemic and considering that IFNλ4 can elicit an antiviral response
against RNA viruses, including some coronaviruses, several studies have assessed whether
rs12979860 is involved in SARS-CoV-2
susceptibility and COVID-19 outcome. In
this regard, it was reported that the T allele
was overexpressed in COVID-19 patients compared to the general healthy population (36.2% vs.
26.4%), thus, this allele was proposed
as a possible risk factor for COVID-19
(Saponi-Cortes et
al., 2021). This was also supported by Rahimi et al. (2021), who demonstrated a positive
correlation between the survival rate in
COVID-19 patients and the rs12979860-CC genotype, which is also favorable to control other
infectious diseases caused by RNA
viruses. On the other hand, a higher
frequency of the CC genotype among COVID-19
patients was reported in a different study, suggesting that people with the C allele (both CT or CC
genotypes) are more susceptible to
SARS-CoV-2 infection (Agwa et al., 2021). However, only slight differences between infected and control groups are shown (44.7%
vs. 44.0%, respectively) and allelic
frequencies are the same for both
groups (C=34.0%, T=66.0%). In that study, it
was also reported that 52.6% of the TT genotypes were classified as severe disease compared to
45.8% and 34.9% in the TC and CC
genotypes, respectively (Agwa et al.,
2021), which seem to be in line with the results published by Saponi-Cortes et al. (2021)
and Rahimi et al. (2021). It is
be noted, also, that the differences shown
by Agwa et al. (2021) may not be exclusively explained by rs12979860 variants, considering that
comorbidities were found in 57.4% of
the infected group (and in 18.0% of
controls). This highlights the relevance of carrying out a properly designed and unbiased sampling as
well as a cautious analysis of the
results in order to discern this type
of controversies when assessing the differential distribution of these variants in different
populations.
CONCLUSIONS
Given its importance and its
apparent association with different
infectious diseases, there is a growing
interest in assessing IFNλ4 polymorphisms. As a whole,
this study describes for the first time the distribution of rs12979860 polymorphism in a healthy sample
of the population of Buenos Aires,
Argentina, further demonstrating that
these frequencies are associated to the
composition of the population. This, in addition to being useful in anthropological studies, may
contribute to the study of different
infectious diseases for which
interferon antiviral responses are key.
ACKNOWLEDGMENTS
We thank Dr. Karina Trono for
critical reading of the manuscript. This research was funded by the services provided
by AC´s group through STAN-CONICET. Other support came from PIP CONICET 2111.
BIBLIOGRAPHY
Agwa S.H.A., Kamel M.M., Elghazaly
H., Abd Elsamee A.M., Hafez H., Girgis S.A.,
Elarab H.E., Ebeid F.S.E., Sayed S.M., Sherif L., Matboli M. (2021) Association between
Interferon-Lambda-3 rs12979860, TLL1
rs17047200 and DDR1 rs4618569 Variant Polymorphisms with the Course and Outcome of SARS-CoV-2 Patients. Genes,
12(6). https://doi.org/10.3390/GENES12060830
Angulo J., Pino K., Echeverría-Chagas
N., Marco C., Martínez-Valdebenito C., Galeno H., Villagra E., Vera L., Lagos N.,
Becerra N., Mora J., Bermúdez A., Cárcamo
M., Díaz J., Miquel J.F., Ferres M., López-Lastra M. (2015) Association of
Single-Nucleotide Polymorphisms in IL28B,
but Not TNF-α, With Severity of
Disease Caused by Andes Virus. Clinical
Infectious Diseases, 61(12): e62-e69. https://doi.org/10.1093/CID/CIV830
Avena S.A., Goicoechea S.A., Rey J.,
Dugoujon J.M., Dejean C.B., Carnese F.R. (2006) Gene mixture in a population
sample from Buenos Aires City. Medicina,
66(2): 113-118.
Avena S., Via M., Ziv E., Pérez-Stable
E.J., Gignoux C.R., Dejean C., Huntsman S.,
Torres-Mejía G., Dutil J., Matta J.L., Beckman K., Burchard E.G., Parolin M.L., Goicoechea A., Acreche
N., Boquet M., Ríos Part M.D.C., Fernández V., Rey J., Fejerman L. (2012) Heterogeneity
in genetic admixture across different
regions of Argentina. PloS One, 7(4).
https://doi.org/10.1371/JOURNAL.PONE.0034695
Bamford C.G.G., Aranday-Cortes E.,
Filipe I.C., Sukumar S., Mair D., Filipe A. da S., Mendoza J.L., Garcia K.C., Fan S., Tishkoff S.A.,
McLauchlan J. (2018) A polymorphic
residue that attenuates the antiviral
potential of interferon lambda 4 in hominid lineages. PLoS Pathogens, 14(10). https://doi.org/10.1371/journal.ppat.1007307
Booth D., George J. (2013) Loss
of function of the new interferon IFN-λ4 may confer protection from
hepatitis C. Nat. Genet. 45(2): 119-120.
https://doi.org/10.1038/NG.2537
Bravo D., Solano C., Giménez E., Remigia
M.J., Corrales I., Amat P., Navarro D. (2014)
Effect of the IL28B Rs12979860 C/T
polymorphism on the incidence and
features of active cytomegalovirus
infection in allogeneic stem cell
transplant patients. J. Med. Virol. 86(5):
838-844. https://doi.org/10.1002/jmv.23865
Chatterjee K. (2010) Host genetic
factors in susceptibility to HIV-1
infection and progression to AIDS. J.
Genet. 89(1): 109-116. https://doi.org/10.1007/s12041-010-0003-4
Chmelova K., Frankova S., Jirsa M.,
Neroldova M., Sticova E., Merta D., Senkerikova R., Trunecka P., Spicak J., Sperl J. (2019) IL28B rs12979860 T allele protects against
CMV disease in liver transplant
recipients in the post-prophylaxis and
late period. Transpl. Infec. Dis. 21(4):
e13124. https://doi.org/10.1111/tid.13124
da Silva Cezar R.D., da Silva
Castanha P.M., Matos Freire N., Mola C.,
Feliciano do Carmo R., Tenório Cordeiro M., Baptista P., Silva Vasconcelos L.R.,
Moura P., da Silva Teixeira V.G. (2020) Association between interferon lambda 3 rs12979860 polymorphism and clinical outcome in dengue
virus-infected children. Int. J.
Immunogenet. 47(4): 351-358. https://doi.org/10.1111/iji.12477
Echeverría N., Chiodi D., López P.,
Sanchez Ciceron A., Angulo J., López-Lastra M., Silvera P., Canavesi A., Bianchi C., Colistro V.,
Cristina J., Hernandez N., Moreno P. (2018) IL28B gene polymorphism rs12979860, but not rs8099917, contributes to the
occurrence of chronic HCV infection in
Uruguayan patients. Virol. J. 15(1): 1-10.
https://doi.org/10.1186/s12985-018-0946-2
Fan W., Xie S., Zhao X., Li N., Chang
C., Li L., Yu G., Chi X., Pan Y., Niu J.,
Zhong J., Sun B. (2016) IFN-λ4 desensitizes the response to
IFN-α treatment in chronic hepatitis C
through long-term induction of USP18. J. Gen. Virol. 97(9): 2210-2220. https://doi.org/10.1099/JGV.0.000522
Fang M.Z., Jackson S.S., O’Brien T.R.
(2020) IFNL4: Notable variants and
associated phenotypes. Gene, 730: 144289.
https://doi.org/10.1016/J.GENE.2019.
Fejerman L., Carnese F.R., Goicoechea
A.S., Avena S.A., Dejean C.B., Ward R.H.
(2005) African ancestry of the
population of Buenos Aires. Am. J.
Phys. Anthropol. 128(1): 164-170. https://doi.org/10.1002/AJPA.20083
Ge D., Fellay J., Thompson A.J., Simon
J.S., Shianna K.V., Urban T.J., Heinzen E.L.,
Qiu P., Bertelsen A.H., Muir A.J., Sulkowski M., McHutchison J.G., Goldstein D.B. (2009) Genetic variation in IL28B predicts
hepatitis C treatment-induced viral
clearance. Nature, 461(7262), 399-401.
https://doi.org/10.1038/nature08309
Hamming O.J., Terczyńska-Dyla
E., Vieyres G., Dijkman R., Jørgensen S.E.,
Akhtar H., Siupka P., Pietschmann T., Thiel V., Hartmann R. (2013) Interferon lambda 4 signals via
the IFNλ receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32(23): 3055-3065. https://doi.org/10.1038/emboj.2013.232
Indolfi G., Mangone G., Calvo P.L.,
Bartolini E., Regoli M., Serranti D., Calitri C., Tovo P.A., De Martino M., Azzari
C., Resti M. (2014a) Interleukin 28B
rs12979860 single-nucleotide
polymorphism predicts spontaneous
clearance of hepatitis C virus in
children. J. Pediatr. Gastroenterol. Nutr.
58(5): 666-668. https://doi.org/10.1097/MPG.0000000000000275
Indolfi G., Mangone G., Bartolini
E., Nebbia G., Calvo P.L., Moriondo M., Tovo P.A., De Martino M., Azzari C., Resti
M. (2014b) Comparative analysis of
rs12979860 SNP of the IFNL3 gene in
children with hepatitis C and ethnic
matched controls using 1000 Genomes Project
data. PloS One, 9(1): e0085899. https://doi.org/10.1371/JOURNAL.PONE.0085899
Ispirologlu M., Bahcecioglu I.H.,
Demirel U., Yalniz M. (2017) Impact of
interleukin 28B rs12979860 C/T
polymorphism on severity of disease and
response to treatment in hepatitis
delta. J. Infect. Dev. Ctries. 11(1): 58-64.
https://doi.org/10.3855/jidc.6872
Key F.M., Peter B., Dennis M.Y., Huerta-Sánchez
E., Tang W., Prokunina-Olsson L., Nielsen R., Andrés A.M. (2014) Selection on a Variant
Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization
of Interferon Lambda 4 (IFNL4). PLoS
Genetics, 10(10): e1004681. https://doi.org/10.1371/journal.pgen.1004681
Kotenko S.V., Gallagher G., Baurin
V.V., Lewis-Antes A., Shen M., Shah N.K., Langer J.A., Sheikh F., Dickensheets H., Donnelly R.P. (2003) IFN-λs mediate antiviral protection
through a distinct class II cytokine receptor complex. Nat Immunol. 4(1): 69-77. https://doi.org/10.1038/ni875
Lazear H.M., Nice T.J., Diamond M.S.
(2015) Interferon-λ: Immune Functions at Barrier
Surfaces and Beyond. Immunity, 43(1): 15-28.
https://doi.org/10.1016/j.immuni.2015.07.001
Machicote A., Flichmann D., Arana
E., Paz S., Fainboim H., Fainboim L., Fernández P. M., Muñiz F.J., Aires B. (2018) IL28B SNPs rs12979860 and rs8099917 Are Associated with Inflammatory Response in Argentine Chronic HCV Patients. Int. J. Clin. Med. 9: 79-
91. https://doi.org/10.4236/ijcm.2018.92009
Mersha T.B., Abebe T. (2015) Self-reported
race/ ethnicity in the age of genomic
research: its potential impact on
understanding health disparities. Hum.
Genomics, 9(1): 1-15. https://doi.org/10.1186/S40246-014-0023-X
Miretti M.M., Beck S. (2006). Immunogenomics: molecular hide and seek. Hum. Genomics,
2(4): 244-251. https://doi.org/10.1186/1479-7364-2-4-244
Muzzio M., Motti J.M.B., Paz
Sepulveda P.B., Yee M., Cooke T., Santos
M.R., Ramallo V., Alfaro E.L., Dipierri J.E.,
Bailliet G., Bravi C.M., Bustamante C.D., Kenny E.E. (2018) Population structure in Argentina. PloS One, 13(5):
e0196325. https://doi.org/10.1371/JOURNAL.PONE.0196325
O’Brien T.R., Prokunina-Olsson L.,
Donnelly R.P. (2014) IFN-λ4: the paradoxical new member of
the interferon lambda family. J.
Interferon Cytokine Res. 34(11): 829-838. https://doi.org/10.1089/JIR.2013.0136
Paschetta C., de Azevedo S., Ramallo
V., Cintas C., Perez O., Navarro P., Bandieri L., Quinto-Sanchez M., Adhikari K.,
Bortolini M.C., Poletti Ferrara G., Gallo C., Bedoya G., Rothhammer F., Acuסa Alonzo V., Ruiz-Linares A., Gonzalez-Jose R. (2021) The impact of socioeconomic and phenotypic traits on self-perception of ethnicity in
Latin America. Sci Rep 11(1):12617. https://doi.org/10.1038/s41598-021-92061-x
Peakall R., Smouse P.E. (2012) GenAlEx
6.5: genetic analysis in Excel.
Population genetic software for teaching
and research--an update. Bioinformatics,
28(19): 2537-2539. https://doi.org/10.1093/BIOINFORMATICS/BTS460
Pedergnana V., Abdel-Hamid M., Guergnon
J., Mohsen A., Le Fouler L., Theodorou I., Mohamed M.K., Fontanet A., Plancoulaine S., Abel
L. (2012) Analysis of IL28B variants in
an Egyptian population defines the 20
Kilobases minimal region involved in
spontaneous clearance of hepatitis C
virus. PLoS ONE, 7(6): e38578. https://doi.org/10.1371/journal.pone.0038578
Peng K., Safonova Y., Shugay M., Popejoy
A.B., Rodriguez O.L., Breden F., Brodin P., Burkhardt A.M., Bustamante C., Cao-Lormeau
V.M., Corcoran M.M., Duffy D., Fuentes-Guajardo
M., Fujita R., Greiff V., Jönsson V.D., Liu
X., Quintana-Murci L., Rossetti M., … Mangul
S. (2021) Diversity in immunogenomics:
the value and the challenge. Nature Methods,
18(6): 588-591. https://doi.org/10.1038/S41592-021-01169-5
Porto L.C., Fabrício-Silva G.M., Poschetzky
B., Perez R., Carneiro dos Santos R., Cavalini
L. (2015) Association of cytokine gene
polymorphisms with hepatitis C virus
infection in a population from Rio de Janeiro, Brazil. Hepat. Med. 7: 71-79. https://doi.org/10.2147/HMER.S89447
Prokunina-Olsson L. (2019) Genetics
of the Human Interferon Lambda Region. J.
Interferon Cytokine Res . 39(10): 599-608. https://doi.org/10.1089/jir.2019.0043
Prokunina-Olsson L., Muchmore B.,
Tang W., Pfeiffer R.M., Park H., Dickensheets H., Hergott D., Porter-Gill P., Mumy A., Kohaar I.,
Chen S., Brand N., Tarway M., Liu L., Sheikh
F., Astemborski J., Bonkovsky H.L., Edlin
B.R., Howell C.D., O’Brien T.R. (2013) A
variant upstream of IFNL3 (IL28B) creating
a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nature Genetics, 45(2): 164-171. https://doi.org/10.1038/ng.2521
Rahimi P., Tarharoudi R., Rahimpour
A., Mosayebi Amroabadi J., Ahmadi I., Anvari
E., Siadat S.D., Aghasadeghi M., Fateh A.
(2021) The association between
interferon lambda 3 and 4 gene
single-nucleotide polymorphisms and the
recovery of COVID-19 patients. Virol. J.
18(1): 1-7. https://doi.org/10.1186/S12985-021-01692-Z
Ramamurthy N., Marchi E., Ansari M.A.,
Pedergnana V., Mclean A., Hudson E., Bowden
R., Spencer C.C.A., Barnes E., Klenerman P. (2018) Impact of Interferon Lambda
4 Genotype on Interferon-Stimulated
Gene Expression During Direct-Acting
Antiviral Therapy for Hepatitis C. Hepatology,
68(3): 859-871. https://doi.org/10.1002/hep.29877
Randolph H.E., Fiege J.K., Thielen
B.K., Mickelson C.K., Shiratori M., Barroso-Batista
J., Langlois R.A., Barreiro L.B. (2021) Genetic ancestry effects on the response to
viral infection are pervasive but cell
type specific. Science, 374(6571): 1127-1133.
https://doi.org/10.1126/SCIENCE.ABG0928
Rizzo S.R.C.P., Gazito D., Pott-Junior
H., Latini F.R.M., Castelo A. (2016) Prevalence of IFNL3 gene polymorphism among blood donors and its relation to genomic profile of
ancestry in Brazil. Braz. J. Infect.
Dis. 20(6): 619-622.
https://doi.org/10.1016/J.BJID.2016.10.002
Saponi-Cortes J.M.R., Rivas M.D.,
Calle-Alonso F., Sanchez J.F., Costo A., Martin C., Zamorano J. (2021) IFNL4 genetic variant can predispose to COVID-19. Sci. Rep. 11(1): 1-4. https://doi.org/10.1038/S41598-021-00747-Z
Sharafi H., Pouryasin A., Alavian
S.M., Behnava B., Keshvari M., Mehrnoush L., Salimi S., Kheradvar O. (2012) Development and validation of a simple, Rapid and
inexpensive PCR-RFLP method for
genotyping of common IL28b
polymorphisms: A useful pharmacogenetic
tool for prediction of Hepatitis C
treatment response. Hepat. Mon. 12(3): 190-195.
https://doi.org/10.5812/hepatmon.849
Sheppard P., Kindsvogel W., Xu W.,
Henderson K., Schlutsmeyer S., Whitmore T.E., Kuestner R., Garrigues U., Birks C.,
Roraback J., Ostrander C., Dong D., Shin
J., Presnell S., Fox B., Haldeman B., Cooper
E., Taft D., Gilbert T., Klucher K.M. (2003)
IL-28, IL- 29 and their class II
cytokine receptor IL- 28R. Nat. Immunol.
4(1): 63-68. https://doi.org/10.1038/ni873
Taheri S., Aygen B., Korkmaz K., Yıldız
O., Zararsız G., Canatan H. (2015)
Characterization of the Interleukin-28B
Gene rs12979860 C/T Polymorphism
in Turkish Chronic Hepatitis C Patients
and Healthy Individuals. Balk. Med. J.
32(2): 147-155.
https://doi.org/10.5152/BALKANMEDJ.2015.15156
Thomas D.L., Thio C.L., Martin M.P.,
Qi Y., Ge D., Ohuigin C., Kidd J., Kidd K., Khakoo S.I., Alexander G., Goedert J.J., Kirk G.D., Donfield
S.M., Rosen H.R., Tobler L.H., Busch M.P., McHutchison J.G., Goldstein D.B., Carrington M.
(2009) Genetic variation in IL28B and
spontaneous clearance of hepatitis C virus. Nature, 461(7265): 798-801. https://doi.org/10.1038/nature08463
Wu L.S., Wang H., Geng X.P. (2012)
Two IL28B polymorphisms are associated
with the treatment response of
different genotypes of hepatitis C in
different racial populations: A
meta-analysis. Exp. Ther. Med. 3(2): 200-206. https://doi.org/10.3892/ETM.2011.385
Zaidane I., Wakrim L., Oulad
Lahsen A., Bensghir R., Chihab H., Jadid
F.Z., El fihry R., Lamdini H., Fayssel N., Marhoum El Filali K., Oudghiri M., Benjelloun
S., Ezzikouri S. (2018) Interleukin 28B
rs12979860 genotype and Human
Immunodeficiency Virus type 1: Susceptibility,
AIDS development and therapeutic outcome.
Hum. Immunol. 79(1): 70-75.
https://doi.org/10.1016/J.HUMIMM.2017.10.011



