Vol. XXXI Issue 2
Article 2
DOI:
10.35407/bag.2020.31.02.02
ARTÍCULOS ORIGINALES
Sequence analysis
suggests positive selection on the bovine prodynorphin gene
Análisis de secuencias genómicas sugieren que
el gen de la prodinorfina está bajo selección positiva en bovinos
Suqueli García M.F.1
Castellote M.A.2
Corva P.M.1 *
1 Facultad de Ciencias Agrarias, Universidad Nacional de Mar del
Plata, Unidad Integrada Balcarce, C.C. 276, 7620 Balcarce, Argentina.
2 Laboratorio de Agrobiotecnologia, EEA Balcarce. Instituto
Nacional de Tecnologia Agropecuaria, Unidad Integrada Balcarce, C.C. 276, 7620 Balcarce,
Argentina.
* Corresponding
author: Pablo Marcelo Corva corva.pablo@inta.gob.ar
ORCID 0000-0002-0660-3115
ABSTRACT
Dynorphin A is an endogenous opioid peptide that is part of the KNDy
system in the hypothalamus of mammals. This peptide acts as an inhibitor of the
GnRH pulse generation, thus regulating the onset of puberty and reproductive
cycles. The PDYN gene encodes the propeptide Prodynorphin, the precursor
of Dynorphin A. Despite its physiological relevance, PDYN has not
emerged as a candidate gene associated with puberty in genomic association
studies conducted in cattle. The present work aimed to search for signatures of
selection on the PDYN gene among cattle breeds. To this, the whole
genome sequences from 57 samples of ten cattle breeds were used. The samples
were grouped based on breed selection history and their productive differences,
particularly in terms of sexual precocity. The population structure was
analyzed using Principal Component Analyses. To evidence recent selection
processes, neutrality tests, such as Tajima’s D and Fu & Li’s F* and D*
were performed in defined functional regions of PDYN. The putative
promoter of PDYN showed a population structure that is in agreement with
the criteria considered to make the groups. In that region, neutrality tests
were consistently negative and resulted in statistically significant for the
dairy breeds. Also, these breeds exhibited less variability in the haplotype
analyses than the others. The results presented here suggest that regulatory
regions of PDYN could be under positive selection, particularly in dairy
breeds.
Key words: Reproduction; KNDy neurons; Dynorphin; Signatures of selection
RESUMEN
Dinorfina A es un péptido opioide
endógeno que forma parte del sistema KNDy en el hipotálamo de mamíferos. Este
péptido actúa como inhibidor de la generación de los pulsos de GnRH, regulando
así el inicio de la pubertad y los ciclos reproductivos. El gen PDYN codifica
el propéptido Prodinorfina, precursor de Dinorfina A. A pesar de su relevancia
fisiológica, PDYN no ha surgido como gen candidato asociado a pubertad
en estudios de asociación genómicos en bovinos. El presente trabajo tuvo como
objetivo buscar huellas de selección en el gen PDYN entre diferentes
razas bovinas. Para alcanzarlo se utilizaron secuencias genómicas de 57
muestras de diez razas bovinas. Las muestras fueron agrupadas considerando la
historia de selección y las diferencias productivas entre razas,
particularmente en términos de precocidad sexual. La estructura poblacional fue
analizada usando análisis de componentes principales. Para evidenciar procesos
de selección recientes se realizaron pruebas de neutralidad, tales como D de
Tajima y F* y D* de Fu & Li, en diferentes regiones funcionales de PDYN.
El promotor putativo de PDYN mostró una estructura poblacional que es
consistente con los criterios usados para agrupar las razas. En esa región, las
pruebas de neutralidad fueron consistentemente negativas y estadísticamente
significativas en las razas lecheras. Además, estas razas también exhibieron
menor variabilidad en los análisis de haplotipos que las demás razas. Los
resultados presentados aquí sugieren que regiones regulatorias de PDYN estarían
bajo selección positiva, particularmente en razas bovinas lecheras.
Palabras clave: Reproducción; Neuronas
KNDy; Dinorfina;
Huellas de selección
Received: 05/05/2020
Revised version received: 07/15/2020
Accepted: 07/24/2020
INTRODUCTION
Reproductive efficiency is one of the most valued aspects of animal
production. In the particular case of beef and dairy cattle,
reproduction-associated traits, especially of females, are included in the
selection objective of the most modern breeds and they have received much
attention in genetic and genomic studies. Starting with puberty and then
cyclically repeated along productive life, the reproductive process in females
requires the release of the gonadotropin-releasing hormone (GnRH) from the
hypothalamus, which in turn is needed for the secretion of gonadotropins from
the anterior pituitary gland (Amstalden and Williams,
2015; Atkins et al.,
2013). Research devoted to understanding the mechanisms underlying the onset
of puberty and the regulation of subsequent reproductive cycles in different
mammal species contributed to building the “KNDy hypothesis” (Smith et al., 2014). According to this
hypothesis three peptides, Kisspeptin, Neurokinin B, and Dynorphin A, are
co-expressed in the same group of neurons in specific regions of the
hypothalamus (Goodman et al.,
2007). These peptides would have the role of a “pulse generator” that drives
GnRH secretion. Neurokinin B and Dynorphin A have stimulatory and inhibitory
effects on Kisspeptin expression, respectively. Moreover, these neurons also
express the Neurokinin B receptor and the Dynorphin A receptor, but not the
Kisspeptin receptor that is expressed in GnRH neurons (Weems et al.,
2018). The regulatory system involving the KNDy neurons would be sensitive
to sex steroid feedback and different regulatory cues (e.g. energy
balance, lactation, photoperiod, stress) but the specific mechanisms involved
are not clear (Lehman et al.,
2010). The role of Dynorphin A in the KNDy neurons is consistent with
previous evidence showing an inhibitory effect of endogenous opioid peptides on
reproduction (Malven, 1986). Suckling increases
levels of endogenous opioid peptides in the brain, making the hypothalamus more
sensitive to the negative feedback from estrogen and decreasing the production
of GnRH (Squires, 2010). Despite the role of
Kisspeptin, Neurokinin B and Dynorphin A in the regulation of the reproductive
cycle, the corresponding genes (KISS1, TAC3, and PDYN) and
their receptors (KISS1R, TACR3, and OPRK1) have not been
analyzed as functional or positional candidates in cattle. The only known
exception is the Dynorphin receptor (OPRK1), proposed as a candidate
gene associated with sexual precocity in a GWAS for sexual precocity in Nellore
cattle from Brazil (Irano et al.,
2016). From the standpoint of animal breeding, the dissection of genetic
variation at the level of the KNDy system could be of help to understand
differences in reproductive performance among cattle breeds, and in turn, to
improve management strategies. As an example, we have evaluated the
reproductive efficiency of Angus and Argentinean Creole females (Corva et al., 1995). The Creole is a
local beef breed that has been much less selected than Angus, but it is
appreciated due to its rusticity and some other favorable traits such as
calving ease. The superiority of Angus over Creole in reproductive performance
was noticed starting with age at puberty (Pardo et al.,
2018) and other differences between the two breeds were detected in
subsequent reproductive cycles (Corva et al., 1995). While body weight
change during the breeding season was a key factor defining pregnancy rate in
Angus females, lactation and the interaction with the offspring was the most
limiting factor in Creole females. The marked difference between European (Bos
taurus) and Indicine (Bos indicus) breeds in sexual precocity is
another well-known example of genetic variation in reproductive performance (Freetly et al., 2011; Laster et al.,
1979) even when were evaluated in similar restrictive environmental
conditions (Meirelles et al.,
1994; Ferraz Jr. et al.,
2018). Taking together the evidences about the inhibitory effect of
endogenous opioid peptides on reproduction and the recently proposed role of
Dynorphin A in KNDy neurons, we hypothesized that PDYN, the gene that
codes for the propeptide Prodynorphin, which is cleaved to produce several
opioid peptides including Dynorphin A (Day et al., 1998, Figure 1), could be under
selection in cattle. This selection probably resulting in the relaxation of the
effects of different cues that convey to the hypothalamus to regulate the onset
of reproductive activity. To test this hypothesis, we compared the sequence of
the PDYN gene from different cattle breeds, taking advantage of the
availability of whole genome sequences.
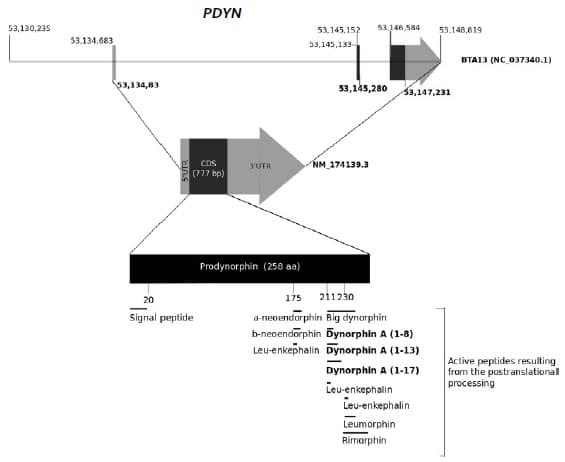
Figure 1. Structure of the bovine
PDYN gene (Top), the corresponding transcript (Genbank sequence:
NM_174139.3) (Middle) and the translated propeptide (Bottom). The different
functional peptides resulting from proteolytic cleavage by the enzyme
Prohormone convertase 2 are also indicated in the figure.
MATERIALS AND METHODS
Retrieval and
processing of PDYN genomic sequences
The genomic sequence of the Bovine Reference Genome (ARS-UCD1.2)
corresponding to a region of BTA13 spanning the PDYN gene (GenBank
accession: NC_037340.1: 53,130,235 - 53,148,819 bp) was downloaded from the
NCBI site (http://www.ncbi.nlm. nih.gov). This DNA sequence was used as a
reference to retrieve the corresponding genomic sequence of Bos indicus (GenBank
accession: NC_032662.1: 53,688,047 - 53,707,634 bp) and also the genomic
sequences of a panel of individuals from selected bovine breeds as it is
described below. The sequenced genomes of selected animals had already been
aligned to the Bovine Reference Genome UMD 3.1.1 (Merchant et
al., 2014; Zimin et al.,
2009). Sequence alignment confirmed that apart from a change in relative
coordinates, the region of interest resulted in identical between UMD 3.1.1 and
ARS-UCD1.2, the latest version of the Bovine Reference Genome
(wwww.bovinegenome.org). The whole panel of genomic sequences used for the
analyses included: The reference Bos taurus and Bos indicus genomes,
corresponding to the Hereford and Nellore breeds respectively; one individual
representing extinct wild cattle (Auroch, Bos primigenius, Park et al., 2015) and individuals from
the “1000 Bull Genomes Project” (http://www.1000bullgenomes.com/) or the U.S.
Meat Animal Research Center (USMARC) Beef Cattle Diversity Panel 2.9
(MBCDPv2.9, Heaton et al.,
2016). Selected breeds were: Holstein (n=13), Jersey (n=11), Angus (n=10),
Limousine (n=6), Corriente (n=4), Longhorn (n=3) and Brahman (n=7). In the
first experiment, the average sequencing depth was 8.3X (Daetwyler et
al., 2014) while in the experiment of Heaton et al.
(2016) it was 14.8X. Aligned genomic sequences (in bam format) were downloaded
from the Sequence Read Archive (SRA) division of NCBI
(http://www.ncbi.nlm.nih.gov/sra) and the USMARC website
(https://www.ars.usda.gov/plainsarea/ clay-center-ne/marc/wgs/bovref/),
respectively. A detailed list of these genomic sequences is presented in Table
1. Based on breed selection history and well known productive differences among
bovine biotypes (particularly in terms of sexual precocity; Diskin and Kenny, 2014; Sartori et al.,
2016) four contrasting groups were defined: highly selected Bos taurus dairy
(Holstein, Jersey, n=24) and beef breeds (Angus, Limousine, Hereford, n=17); less
selected -hardy- Bos taurus breeds (Corriente, Longhorn, Auroch, n=8)
and zebu breeds (Brahman, Nellore, n=8). These groups are hereinafter
identified as “Dairy”, “Beef”, “Less selected” and “Zebu”, respectively. Each
genomic sequence in bam format was searched for variant sites using BCFtools
software v1.5 (Li, 2011) with the command: “bcftools mpileup -f sequence.fasta
input.bam | bcftools call -m -Ob | tabix | bcftools query -f ‘%POS\t%QUAL\t%ALT[\t%IUPACGT]\n’
-o output. txt” where “sequence.fasta” was the sequence of the Bovine Reference
Genome UMD 3.1.1 used as a reference in the original alignment. Variants with a
Phred quality score below 20 (base call accuracy above 99%) were manually
discarded and a consensus sequence was obtained for each sample in fasta format
using UNIX commands. The average depth of coverage of each sample for the whole
genome sequence was retrieved from the original database. On the other hand,
the average depth of coverage of each sample for the PDYN gene region was
calculated using Mosdepth software v 0.2.9 (Pedersen and
Quinlan, 2018) (Table 1).
Table 1. Genomic sequences used
to characterize the bovine Prodynorphin gene. Each retrieved sequence was
aligned to the region of BTA13 spanning the PDYN gene (Bovine Reference
Genome ARS-UCD1.2, GenBank accession: NC_037340.1: 53,129,235-53,148,819).
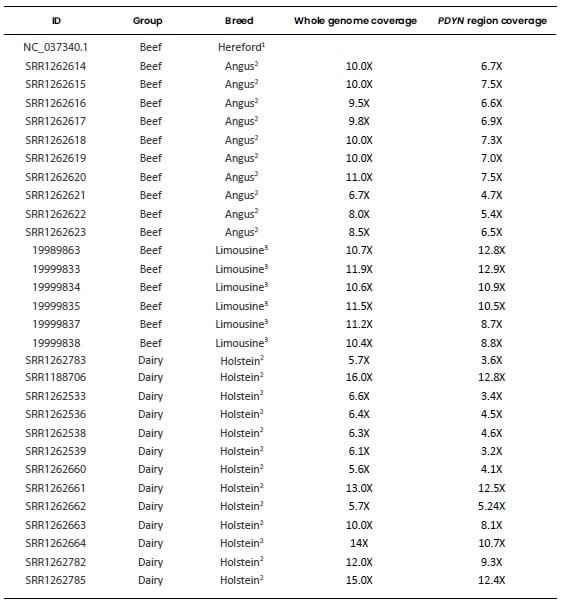
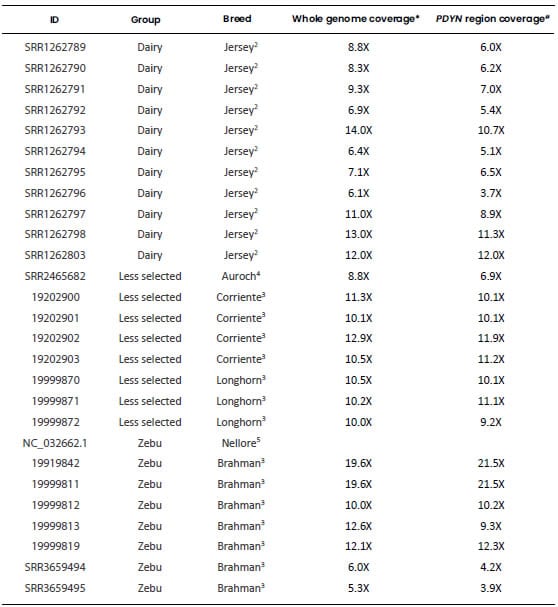
ID: File identification
in the original database. Dairy: Highly selected dairy breeds (Bos taurus);
Beef: Highly selected beef breeds (Bos taurus); Less selected: less
selected breeds (Bos taurus), includes the Auroch (Bos primigenius);
Zebu: zebu breeds (Bos indicus). All samples were aligned to the genomic
region of BTA13 spanning the PDYN gene and only this region of each
sample was used for the analyses. 1Bovine Reference Genome ARS-UCD1.2, GenBank
accession: NC_037340.1: 53,130,235-53,148,819). 2“1000 Bull Genome Project” Run
2, (http://www.1000bullgenomes.com/). 3U.S. Meat Animal Research Center
(USMARC) Beef Cattle Diversity Panel 2.9 (MBCDPv2.9, Heaton et al.,
2016). 4Bos primigenius archeological sample (Park et al., 2015). 5Bos indicus Reference
Genome Bos_indicus_1.0 (GenBank accession: NC_032662.1). *Average depth of
coverage obtained from their respective original database. #Average depth of
coverage calculated by using Mosdepth software v 0.2.9 (Pedersen and
Quinlan, 2018).
Haplotype
reconstruction
The Prodynorphin cDNA was cloned by Jiang et al. (1997); Genbank accession
U58500.1. This sequence agrees with accession NM_174139.3 from the annotation
of the last version of the Bovine Reference Genome (ARSUCD1.2), and therefore
it was used in the present work as a reference of gene organization. In the
comparative sequence analyses, the whole sequence and four functional regions
of the mRNA were considered: a 1,000 bp fragment spanning the putative proximal
promoter; the 5’ untranslated region (5’UTR, 174 bp); the coding sequence (CDS,
777 bp); and the 3’ untranslated region (3’UTR, 1,588 bp) (Figure 1). The sequences of the breed panel were aligned
online with the MAFFT software version 7
(https://mafft.cbrc.jp/alignment/server/) and the resulting alignment was
manually edited using the Aliview software version 1.23 (Larsson, 2014). Then, haplotype
phases from sequences with heterozygous sites were reconstructed with DnaSP
version 6.10.03 (Rozas et al.,
2017) using the option fastPHASE (Scheet and Stephens,
2006) with default parameters. Haplotypes were reconstructed for the whole
sequence and then trimmed into the functional regions as defined above. The
phylogenetic relationships among the haplotypes identified in the putative
proximal promoter (1,000 bp) and in the coding sequence (CDS) of the PDYN gene,
were inferred through a median-joining network analysis (Bandelt et al.,
1999) using the PopART software version 1.7 (Leigh and Bryant,
2015).
Principal component
analyses (PCA)
To evaluate population structure and the genetic relationships among the
four breed groups (“Dairy”, “Beef”, “Less selected” and “Zebu”), principal
component analyses (PCA) were computed on haplotype frequencies of both, the
putative proximal promoter region (1,000 bp) and the cDNA (complementary DNA,
2,539 bp), of PDYN using the ‘prcomp’ function of Rstudio v.3.4.4 (R
Development Core Team, 2018). PCA results were plotted using the R package
‘ggplot2’ (Wickham, 2016).
Signatures of selection
To detect departures from the expectations of the neutral theory of
evolution (Kimura, 1968) the neutrality tests Tajima’s
D (Tajima, 1989) and Fu & Li’s F* and D* (Fu
and Li, 1993) were conducted. Tajima’s D compares the estimate of DNA sequence variation
based on the average pairwise distance between all sequences in the sample (φ), to the estimate of DNA sequence
variation based on the observed number of segregating sites and the number of
chromosomes in a sample (θ). Under neutrality,
the means θ and φ should be approximately equal to each
other. Therefore, the expected value of Tajima’s D for populations conforming
to a standard neutral model is zero. Significant deviations from zero indicate
a skew in the allele frequency distribution relative to neutral expectations.
Positive values of Tajima’s D arise from an excess of intermediate frequency
alleles and can result from population bottlenecks, structure and/ or balancing
selection. Negative values of Tajima’s D indicate an excess of low frequency
alleles and can result from population expansions or positive selection
(Tajima, 1989; Biswas and Akey, 2006). On the other hand,
Fu & Li´s test makes the distinction between old and recent mutations as
determined by where they occur on the branches of genealogies. The D* and F*
statistics compare an estimate of the population mutation rate based on the
number of derived variants seen only once in a sample (referred to as
singletons) with φ or θ, respectively. Fu & Li´s F* and D*
values are negative when there is an excess of recent mutations and will be
taken as evidence against the neutrality of mutations (Fu and Li, 1993; Biswas
and Akey, 2006). These tests were conducted in the four defined cattle groups
for the whole PDYN gene and also for each functional region defined
above, using DnaSP version 6.10.03 (Rozas et al.,
2017).
RESULTS
Principal component
analyses
The PCA conducted on the haplotype frequencies of the cDNA and the
putative promoter of PDYN are presented in Figure 2. It can be seen that
the distribution of the breed groups does not agree between both gene regions.
In the case of the promoter, PC1 alone explains 79.7% of the variability.
Breeds are distributed along this axis, in agreement with the criteria that
were originally considered to make the groups (selection history and,
particularly, sexual precocity) suggesting a population structure defined not
only by breed isolation and genetic drift but probably also by selection. On the
contrary, when haplotype frequencies of the cDNA are considered, PC1 and PC2
explain 37.1% and 32.4% of the variation, respectively, and breed distribution
do not match grouping criteria.
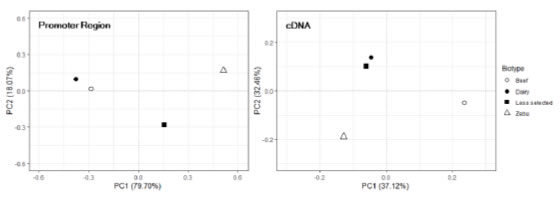
Figure 2. Bidimensional plot
illustrating the results of the Principal Component Analysis performed on
haplotype frequencies of the putative proximal promoter (left) and the cDNA
(NM_174139.3) (right) of the bovine PDYN gene. The percentages of total
variability explained by each PC are included between brackets. Dairy: Highly
selected dairy breeds (Bos taurus); Beef: Highly selected beef breeds (Bos
taurus); Less selected: Less selected breeds (Bos taurus), included
the Auroch (Bos primigenius); Zebu: Zebu breeds (Bos indicus).
Signatures of selection
Departures from neutral theory expectations were analyzed on each of the
four defined breed groups (“Dairy”, “Beef”, “Less selected” and “Zebu”) (Table
4). In the “Dairy” cattle group, these tests were consistently negative and
resulted to be statistically significant for the Genomic sequence as well as
for the putative promoter region (Table 2). These results suggest that this
gene and particularly the promoter region could be under positive selection.
Moreover, these results pointed to a potential effect on regulatory features of
PDYN gene expression in Dairy cattle. The other groups, on the contrary,
had much higher nucleotide and haplotype diversity in the promoter and CDS than
the Dairy breeds, and none of their neutrality tests were statistically
significant (Table 2).
Table 2. Diversity measures and
neutrality tests. Results corresponding to the whole genomic sequence of PDYN
and each of the different functional regions of transcript NM_174139.3 are presented
for each group of breeds.
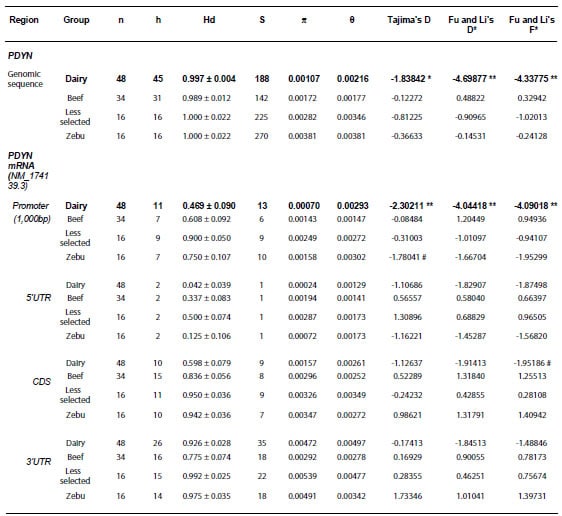
n: number of
chromosomes; h: number of haplotypes; Hd: haplotype diversity; S: number of
segregating sites; π: nucleotide diversity
(per site); θ= 4Nμ (N and μ are the effective population size, and the
mutation rate per DNA sequence per generation, respectively); Dairy: Highly
selected dairy breeds (Bos taurus); Beef: Highly selected beef breeds (Bos
taurus); Less selected: less selected breeds (Bos taurus), includes
the Auroch (Bos primigenius); Zebu: zebu breeds (Bos indicus).
#0.1>p>0.05; *p<0.05; **p<0.01. A region of the gene was considered
under selection when both Tajima’s and Fu & Li’s neutrality tests produced
significant results. These regions are shown in bold. 5’UTR and 3’UTR:
untranslated regions; CDS: coding sequence.
Haplotype description
Table 3 shows the mutations identified in the promoter region of the PDYN
gene that had evidence of positive selection according to the neutrality
tests, and in the CDS. These mutations define the haplotypes reported in Table
4. The combination of 14 SNPs defined the haplotypes of the promoter region
(1,000 bp) (Table 4). In this region, a total of 32 haplotypes were found among
the 114 chromosomes (57 samples by 2 chromosomes each) analyzed. Sixteen of
them were found in at least two chromosomes and were represented by 98
chromosomes (Table 4). The most abundant haplotype (p-Hap_1, ref; n=56) was
found in the Bovine Reference Genome ARSUCD1.2 but it was absent in the “Zebu”
cattle group. In the CDS (777 bp), a total of 28 haplotypes defined by 10 SNPs
were found among the 114 chromosomes analyzed. Twelve of them were found in at
least two chromosomes and were represented by 98 chromosomes (Table 3). In this
region, the haplotype found in the Bovine Reference Genome ARS-UCD1.2 (c-Hap_1,
ref; n=15) was not the most abundant and was the only one with the “A” allele
in the SNP rs42388967 (SNP 3 in the CDS, Table 4). Although “Beef” and “Dairy”
groups shared their most frequent haplotype (p-Hap_9) in the promoter that was
not the case for the CDS (c-Hap_9 and c-Hap_1 for “Dairy” and “Beef” groups,
respectively). The promoter of PDYN had five different haplotypes in
dairy breeds but with little variability, given that 34 out of 40 chromosomes
shared p-Hap_1. This same haplotype was present in 20 out of 33 chromosomes of
selected beef breeds, but the rest of the chromosomes of this group had seven
different haplotypes in low frequencies each (Table 4). Moreover, in the group
of dairy breeds, 40 out of 44 haplotypes of de CDS share the same six alleles
in their 5’ end. This same trend was detected in the promoter, where 37 out of
40 haplotypes share the same seven alleles in their 3’ end. This pattern is not
seen in any other breed group (Table 4). Zebu breeds had four haplotypes with a
predominance of p-Hap_11 in the promoter region and showed little coincidence
in haplotype distribution with the other breed groups (Table 4). Interestingly,
differences between breed groups were smoothed when the CDS was considered and
even the “Zebu” group showed more coincidence with the other Bos taurus groups.
Nevertheless, for the CDS the “Dairy” group showed again the lowest variability
(30 out of 44 chromosomes had Hap_9; Table 4). The “Median Joining” network
constructed using the haplotypes identified in the promoter region suggested
the existence of two clades separated by three mutational steps (Figure 3A). In this network, the haplotypes found in the
“Dairy” cattle group could be differentiated from those of the “Zebu” cattle
group. Noteworthy, the “Dairy” group exhibited few haplotypes and they formed a
star-like configuration with the reference haplotype (p-Hap_1) as central
haplotype in the clade (Figure 3A). In contrast, the
haplotypes from the other groups were sparsely distributed. On the other hand,
the “Median Joining” network constructed using the haplotypes identified in the
CDS did not exhibit a clear pattern of variation and contrary to what happened
in the promoter region, the haplotypes found in the CDS in the different cattle
groups were not separated in this network (Figure 3B). The 28 haplotypes
identified in the CDS (Table 4) could be translated into six propeptide
sequences, which had no more than four changes compared to the translated
reference sequence. The four breed groups coincidentally had the same
translated sequence (translated sequence of c-Hap_9) as the most abundant (Table
4). Interestingly, none of the aminoacid substitutions affected the sequence of
the functional peptides that are processed from the translated propeptide (Table
3, Figure 1).
Table 3. List of mutations
identified in two regions of the PDYN gene (promoter and coding
sequence, CDS).
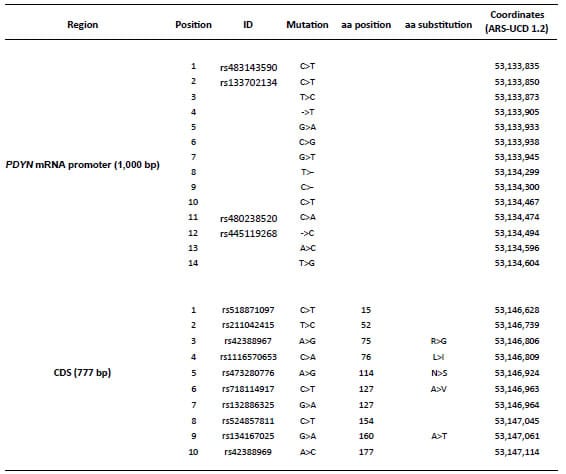
The “Position” column
indicates the position in the corresponding haplotypes described in Table 4.
Table 4. List of haplotypes.
List of haplotypes defined by mutations identified in the coding sequence (CDS)
and in the promoter region of the PDYN. Only haplotypes found in at
least two chromosomes are shown.
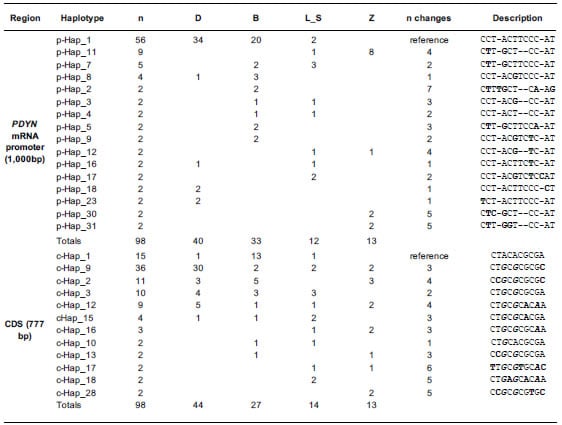
The total number of
identified chromosomes (n) are discriminated by breed group. Dairy: Highly
selected dairy breeds (Bos taurus); Beef: Highly selected beef breeds (Bos
taurus); Less selected: less selected breeds (Bos taurus), includes
the Auroch (Bos primigenius); Zebu: zebu breeds (Bos indicus). n
changes: number of changes respect to the bovine reference genome ARS-UCD1.2.
Differences respect to the reference genome are shown in bold. The positions of
SNPs that generate aa substitutions in the CDS are in bold italic font.
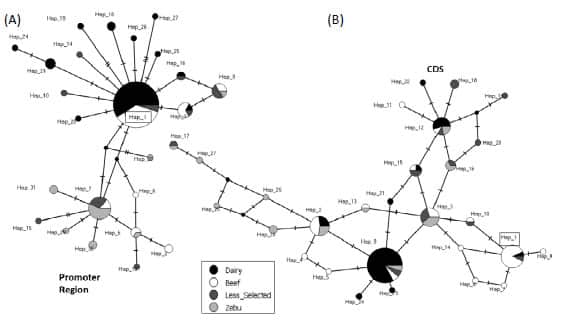
Figure 3. Median Joining network
constructed with the haplotypes corresponding to (A) the putative proximal
promoter (1,000 bp) and (B) the coding sequence (CDS) of PDYN gene. The
size of each circle is proportional to the corresponding chromosome frequency.
Slashes represent the number of mutations separating each haplotype. In each
network Hap_1 (box) corresponds to the Bovine Reference Genome ARS-UCD1.2.
Dairy: Highly selected dairy breeds (Bos taurus); Beef: Highly selected
beef breeds (Bos taurus); Less selected: Less selected breeds (Bos
taurus), included the Auroch (Bos primigenius); Zebu: Zebu breeds (Bos
indicus).
DISCUSSION
The involvement of Endogenous Opioid Peptides (EOP) as negative
regulators of the reproductive cycle has been known for a long time, but their
role has not been completely elucidated. In cattle, EOP received attention as
factors consistently involved in the inhibitory effects of suckling on reproductive
activity (Malven et al.,
1986). Frequent suckling or milking increases levels of EOP in the brain,
which in turn increases the sensitivity of the hypothalamus to the negative feedback
from estrogen (Squires, 2010). This experimental
evidence confirms the role of Dynorphin A and EOP in general, as inhibitors of
reproductive activity in mammals, and justify the search of selection
signatures conducted in the present work to explain the well-known variability
in reproductive performance among cattle breeds, that is noticeable since the
onset of puberty (Diskin and Kenny, 2014). In the present
experiment, we analyzed an annotated sequence of PDYN, which encoded,
among others, the endogenous opioid peptide Dynorphin A (Figure 1), from the last
version of the Bovine Reference Genome (Genbank accession NM_174139.3). This
sequence has a genomic organization that is consistent with the cDNA originally
cloned by Jiang et al. (1997); Genbank accession
U58500.1). The dataset analyzed here included the only available sample from
the Auroch (Bos primigenius, Park et al.,
2015) that were grouped with less selected breeds. This sample did not show
marked differences with other groups. However, it must be noted that a single
sample is not enough to identify signatures of selection. Thus, more genomes
from this group should be sequenced to be more conclusive about the effects of
domestication (Orlando, 2015). The PCA results
presented in Figure 2, showing sample
substructure differed depending in the analyzed regions of PDYN (promoter
and cDNA) indicates that the observed differences are not only justified by the
well-known genetic distance between Bos indicus and Bos taurus breeds,
product of sub-speciation, and justify the definition of four separated groups
used in the other analyses. The comparison of haplotype frequencies among breed
groups for both genomic regions (Table 4, Figure 3) also supports the
existence of directional selection on regulatory regions of the gene and not
only random genetic drift associated with the breed or subspecies formation in
the definition of sequence variation. It could be argued that low genetic
variability is the result of reduced effective population size (Ne), a feature
common to selected populations in most livestock species (Taylor et al.,
2016). In this case, the group of dairy breeds was integrated by two
different breeds, Holstein and Jersey that share many selection objectives and
also a common production system. Also, this group had the largest sample size.
The methods that were used in the present work are appropriate to identify
recent selection (Biswas and Akey, 2006). Indeed, the results
of the Tajima’s D (Tajima, 1989) and Fu & Li D* and F* (Fu and Li, 1993) exhibited statistically
significant negative values in the “Dairy” group (Table 2) which pointed out to
a recent selection process in modern, highly specialized breeds. This evidence
of recent selection is consistent with the emphasis of intense artificial
selection on production traits, particularly those related to reproductive
efficiency. A compelling argument in favor of positive selection on a gene such
as PDYN comes from the intrinsic characteristics of different livestock
production systems. Under extensive conditions, the cow not only nurses but
also protects the progeny, and the maternal bond exerts a strong inhibiting
effect on reproductive activity (Williams, 1990). In more intensified
systems such as those for dairy production the calf is separated from the
mother hours after parturition. Moreover, dairy cows are usually expected to
get pregnant under a strongly negative energy balance (Butler and Smith,
1989; Beam and Butler, 1999; Vercouteren et
al., 2015). Clearly, in these extreme cases, the physiological
and environmental cues that have to be processed at the hypothalamic level to
regulate reproductive cycles are very different. It could be hypothesized that
in females from more intensified production systems, the effects of some
inhibitory mechanisms acting on the regulation of reproduction are relaxed,
favoring the selection response for reproductive efficiency. The results
reported here showed that the “Dairy” group presented fewer haplotypes than the
other cattle groups in the promoter region and that these haplotypes exhibit
low differences among them (Table 4, Figure 3A). A detailed
description on the epigenetic regulation of the expression on KISS1 and TAC3
by implementing histone modifications was recently made by Toro et al. (2018). However, it does not
seem to be the case of PDYN gene, the third member of the triad of the
“KNDy hypothesis” (Smith et al., 2014). The results presented here
warrants further investigation on the mechanisms underlying the PDYN transcriptional
regulation. The results presented here suggest that there is selection pressure
acting on putative regulatory regions on the proximal promoter of PDYN (Table
2), which seems to be a common feature of selection for quantitative or complex
traits. Modifications of the coding sequence are more common in the case of
genes underlying mendelian traits, in which the mutation has a strong effect on
the phenotype (Boyle et al.,
2017). Although aminoacid substitutions on the propeptide were detected in
the present analysis, they do not seem to affect the sequence of functional
peptides, stressing their biological importance (Table 3, Figure 1). Also, a not clear
pattern of differentiation was observed among the haplotypes of the four cattle
groups in the coding sequence (Figure 3B). Selection on
regulatory regions of PDYN would affect the expression of all the
different peptides coded by the gene. Given the physiological role of Dynorphin
A, the interest to improve reproductive efficiency in highly specialized cattle
breeds provides a very reasonable explanation to justify selection on PDYN.
Nevertheless, at this point, the relevance of the other peptides cannot be
ruled out. Further research should clarify the role of EOP, particularly
Dynorphin A, in the regulation of reproduction both within and between cattle
breeds. The usefulness of QTN identification (Quantitative Trait Nucleotides underlying
Quantitative Trait Loci) in animal breeding is a matter of debate. Present
genomic selection in cattle is mostly based on dense arrays of anonymous
markers; however, methods are being developed to include QTN information to
improve prediction accuracy (Fragomeni et al.,
2017). Besides, there are initiatives to enhance progress in animal
selection trough the combination of conventional breeding and gene editing (e.g.:
Promotion of Alleles by Genome Editing; Jenko et al.,
2015). However, there are doubts about the efficacy of this approach due to
the intrinsic polygenic basis of quantitative variation (Simianer, 2018). Independently of
potential direct applications of the knowledge about PDYN variation
among cattle breeds, the present work contributed with elements for a deeper understanding
of the complex interaction among genetic variation, artificial selection, and
environmental effects.
BIBLIOGRAPHY
Amstalden M., Williams G.L. (2015) Neuroendocrine Control of Estrus and
Ovulation. In: Hopper R.M. (Ed.) Bovine Reproduction. Wiley Blackwell, Starkville,
Mississippi, USA, pp. 203-219.
Atkins J.A., Pohler K.G., Smith M.F. (2013) Physiology and Endocrinology
of Puberty in Heifers. Vet. Clin. North. Am. Food. Anim. Pract. 29: 479-492.
Bandelt H.J., Forster P., Röhl A. (1999) Median-Joining Networks for
Inferring Intraspecific Phylogenies. Mol. Biol. Evol. 16: 37-48.
Beam S.W., Butler W.R. (1999) Effects of energy balance on follicular
development and first ovulation in postpartum dairy cows. J. Reprod. Fertil.
Suppl. 54: 411-424.
Biswas S., Akey J.M. (2006) Genomic insights into positive selection. Trends
Genet. 22: 437-446.
Boyle E.A., Li Y.I., Pritchard J.K. (2017) An expanded view of complex
traits: from polygenic to omnigenic. Cell. 169: 1177-1186.
Butler W.R., Smith R.D. (1989) Interrelationships Between Energy Balance
and Postpartum Reproductive Function in Dairy Cattle. J. Dairy Sci. 72: 767-783.
Corva P.M., Villarreal E.L., Mezzadra C.A. (1995) Reproductive traits of
Angus, Criollo and reciprocal crossbred females in the temperate area of
Argentina. Anim. Sci. 61: 241-249.
Daetwyler H., Capitan A., Pausch H. (2014) Whole-genome sequencing of
234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat.
Genet. 46: 858-865.
Day R., Lazure C., Basak A. (1998) Prodynorphin Processing by Proprotein
Convertase 2. J. Biol. Chem. 273: 829-836.
Diskin M.G., Kenny D.A. (2014) Optimising reproductive performance of
beef cows and replacement heifers. Animal 8: 27-39.
Ferraz Jr. M.V.C., Pires A.V., Santos M.H. (2018) A combination of
nutrition and genetics is able to reduce age at puberty in Nelore heifers to
below 18 months. Animal 12: 569-574.
Fragomeni B.O., Lourenco D.A.L., Masuda Y. (2017) Incorporation of
causative quantitative trait nucleotides in single-step GBLUP. Genet. Sel. Evol.
49: 59.
Freetly H.C., Kuehn L.A., Cundiff L.V. (2011) Growth curves of crossbred
cows sired by Hereford, Angus, Belgian Blue, Brahman, Boran, and Tuli bulls,
and the fraction of mature body weight and height at puberty. J. Anim. Sci. 89:
2373-2379.
Fu Y.X., Li W.H. (1993) Statistical tests of neutrality of mutations. Genetics
133: 693-709.
Goodman R.L., Lehman M.N., Smith J.T. (2007) Kisspeptin neurons in the
arcuate nucleus of the ewe express both dynorphin A and neurokinin B. Endocrinology
148: 5752-5760.
Heaton M.P., Smith T.P.L., Carnahan J.K. (2016) Using diverse U.S. beef
cattle genomes to identify missense mutations in EPAS1, a gene associated with
high-altitude pulmonary hypertension. F1000Research 5: 2003.
Irano N., De Camargo G.M.F., Costa R.B. (2016) Genome-wide association
study for indicator traits of sexual precocity in Nellore cattle. PLoS One 11: 1-14.
Jenko J., Gorjanc G., Cleveland M.A. (2015) Potential of promotion of
alleles by genome editing to improve quantitative traits in livestock breeding
programs. Genet. Sel. Evol. 47: 55.
Jiang H., Weesner G.D., Malven P.V. (1997) cDNA sequence and expression
of bovine prodynorphin. Gene 186: 279-283.
Kimura M. (1968) Evolutionary Rate at the Molecular Level. Nature 217: 624-626.
Larsson A. (2014) AliView: a fast and lightweight alignment viewer and
editor for large datasets. Bioinformatics 30: 3276-3278.
Laster D.B., Smith G.M., Cundiff L.V. (1979) Characterization of
biological types of cattle (Cycle II). II Postweaning growth and puberty of
heifers. J.Anim. Sci. 48: 500-508.
Lehman M.N., Coolen L.M., Goodman R.L. (2010) Minireview: Kisspeptin/
neurokinin B/dynorphin (KNDy) cells of the arcuate nucleus: A central node in
the control of gonadotropin-releasing hormone secretion. Endocrinology 151: 3479-3489.
Leigh J., Bryant D. (2015) PopART: Full-feature software for haplotype
network construction. Methods Ecol. Evol. 6: 1110-1116. Available at: http://popart.otago.ac.nz.
Li H. (2011) A statistical framework for SNP calling, mutation
discovery, association mapping and population genetical parameter estimation
from sequencing data. Bioinformatics 27: 2987-2993.
Malven P.V. (1986) Inhibition of pituitary LH release resulting from
endogenous opioid peptides. Domest. Anim. Endocrinol. 3: 135-144.
Malven P.V., Parfet J.R., Gregg D.W. (1986) Relationships among
Concentrations of Four Opioid Neuropeptides and Luteinizing Hormone-Releasing
Hormone in Neural Tissues of Beef Cows Following Early Weaning. J. Anim. Sci. 62: 723-733.
Meirelles C.F., Abdalla A.L., Vitti D.M.S.S. (1994) The effect of feed
supplementation on the onset of puberty in Brazilian dairy heifers. Sci. Agric.
51: 374-380.
Merchant S., Wood D.E., Salzberg S.L. (2014) Unexpected cross-species
contamination in genome sequencing projects. Peer J. 2: e675.
Orlando L. (2015) The first aurochs genome reveals the breeding history
of British and European cattle. Genome Biol. 16: 225.
Pardo A.M., Villarreal E.L., Papaleo Mazzucco J. (2018) Sexual precocity
and productivity of beef cattle female under grazing conditions. Anim. Prod.
Sci. 59: 757-766.
Park S.D.E., Magee D.A., McGettigan P.A. (2015) Genome sequencing of the
extinct Eurasian wild aurochs, Bos primigenius, illuminates the
phylogeography and evolution of cattle. Genome Biol. 16: 234.
Pedersen B.S., Quinlan A.R. (2018) Mosdepth: quick coverage calculation
for genomes and exomes. Bioinformatics 34: 867-868.
R Development Core Team (2018) R: a language and environment for
statistical computing Vienna, Austria: The R Foundation for Statistical Computing.
Available at: http://www.r-project.org/.
Rozas J., Ferrer-Mata A., Sanchez DelBarrio J.C. (2017) DnaSP 6: DNA
sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 34:
3299-3302.
Sartori R., Gimenes L.U., Monteiro Jr. P.L.J. (2016) Metabolic and
endocrine differences between Bos taurus and Bos indicus females
that impact the interaction of nutrition with reproduction. Theriogenology 86: 32-40.
Scheet P., Stephens M. (2006) A Fast and Flexible Statistical Model for
Large-Scale Population Genotype Data: Applications to Inferring Missing
Genotypes and Haplotypic Phase. Am. J. Hum. Genet. 78: 629-644.
Simianer H. (2018) Of Cows and Cars. J. Anim. Breed. Genet. 135: 249-250.
Smith M.F., Hawken P.A.R., Lehman M.N. (2014) The role of kisspeptin in
reproductive function in the ewe. In: Juengel J.L., Miyamoto A., Price C., Reynolds
L.P., Smith M.F., Web R. (Eds.) Reproduction in Domestic Ruminants VIII. Context,
UK, pp. 105-116.
Squires E.J. (2010) Applied Animal Endocrinology. 2nd Ed. CABI
Publishing, Guelph, Ontario, Canada.
Tajima F. (1989) Statistical Method for Testing the Neutral Mutation. Genetics
123: 585-595.
Taylor J.F., Taylor K.H., Decker J.E. (2016) Holsteins are the genomic
selection poster cows. Proc. Natl. Acad. Sci. 113: 7690-7692.
Toro C.A., Wright H., Aylwin C.F. (2018) Trithorax dependent changes in
chromatin landscape at enhancer and promoter regions drive female puberty. Nat.
Commun. 9: 57.
Vercouteren M.M.A.A., Bittar J.H.J., Pinedo P.J. (2015) Factors
associated with early cyclicity in postpartum dairy cows. J. Dairy Sci. 98: 229-239.
Weems P.W., Lehman M.N., Coolen L.M. (2018) The Roles of Neurokinins and
Endogenous Opioid Peptides in Control of Pulsatile LH Secretion. In: Litwack G.
(Ed.) Vitamins and Hormones. Elsevier Inc., Academic Press, pp. 89-135.
Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag,
New York.
Williams G.L. (1990) Suckling as a regulator of postpartum rebreeding in
cattle: a review. J. Anim. Sci. 68: 831-852.
Zimin A.V., Delcher A.L., Florea L. (2009) A whole-genome assembly of
the domestic cow, Bos taurus. Genome Biol. 10: R42.



